Whole genome sequencing of recurrent tuberculosis in Stockholm County 1996–2016
Introduction
Over the last two decades considerable improvement has been made in tuberculosis (TB) diagnosis, treatment and control worldwide. Still in 2017, 10 million people fell ill with TB, according to The World Health Organization (WHO) (1). WHO estimates that the treatment success rate for the 5.9 million new TB cases in the 2016 global cohort was 82% (1). The WHO has set a goal to reduce the TB incidence rate with 90% from 2015 to 2035. To achieve this target all countries should aim to reach a 90% treatment coverage and a 90% treatment success rate (1).
Sweden is a low burden country for TB with a median incidence of 6.0 per 100,000 population 1996–2016 (2). The majority of cases are seen in immigrants from TB endemic areas (2).
Recurrent TB is defined as an episode of TB occurring in a person after treatment completion and can be caused either by endogenous infection with the same strain, relapse, or by reinfection with a different strain (3-5).
Male sex, low socioeconomic status, origin from a high endemic region, diabetes mellitus, smoking, alcohol abuse, intravenous drug use, Beijing lineage strain, a multidrug resistant (MDR) strain, pulmonary cavitation and HIV coinfection have all been shown to be associated with increased risk of TB reoccurrence in TB low endemic regions (6,7).
In patients with recurrent TB, genotyping of the Mycobacterium tuberculosis (M.tb) strains has been suggested as a tool to separate relapse cases from reinfection cases. Based on findings with earlier genotyping methods [restriction fragment length polymorphism (RFLP) typing or mycobacterial interspersed repetitive unit-variable number of tandem repeat (MIRU-VNTR)] reinfection has been considered as the main cause for recurrent infection in high endemic countries (8,9). In contrast relapse has been considered as the main cause of recurrence in low endemic countries using the same methods (6). Whole genome sequencing (WGS) can distinguish relapse from reinfection with higher resolution than the earlier used methods (10). WGS is also a rapid and precise method for genotypic drug resistance analysis with high concordance (95–96%) with culture-based phenotypic drug susceptibility testing methods (11-13).
The aim of this study was to analyze the frequency of TB-recurrence in Stockholm Country and to use WGS to separate recurrent cases caused by relapse from those caused by reinfection as an evaluation of current treatment strategies and disease control measures.
We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/jphe-20-10).
Methods
This was a population-based cohort study conducted by the Karolinska University Hospital in Stockholm, the Public Health Agency of Sweden and the Department of Communicable Disease Control and Prevention at Stockholm County Council in collaboration.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Ethical approval for the study was obtained from the Regional Ethical Review Board in Stockholm (Dnr 2018/1171-31/1). Informed consent was not obtained as no individuals could be identified through the presented data and as a requirement of informed consent would have introduced uneven response bias in the setting of this population based study.
Setting
In Sweden it is mandatory to report TB-cases in the web-based, National reporting system and registry for communicable diseases (SmiNet), according to the Communicable Disease Act. Patients diagnosed with TB in Stockholm are treated exclusively at the Karolinska University Hospital and according to national and international guidelines. Direct observed treatment (DOT) was not routinely used but drugs were distributed to the patients in a dosing box for 2 weeks’ use and refilled at the TB clinic. Samples for TB-diagnosis were analyzed at the laboratory for microbiology at Karolinska University Hospital. All M.tb strains from TB-patients are sent, for molecular epidemiological typing and storage to the Public Health Agency of Sweden.
TB patients are obliged by law to attend the clinic during treatment and at follow-up scheduled at 6 and 12 months after treatment completion.
Data collection
All TB-cases in Stockholm County, from January 1996 (start of SmiNet-registry) through December 2016, were identified in the SmiNet-registry. Only cases with culture confirmed M.tb infection were eligible for inclusion and followed for TB recurrence until the end of 2017. The unique Swedish personal identity number was used for identification. Demographic and clinical data and microbiological results were extracted from the SmiNet-registry. Eventual TB recurrence within the cohort, in other parts of Sweden was controlled for in the same registry. On patients with recurrent TB, information on risk factors for recurrence was retrieved from individual patients files while information regarding HIV-coinfection was retained from the Swedish national quality registry (InfCare HIV) which includes more than 99% of residents living with HIV in Sweden since 1983 (14).
WGS was used to distinguish relapse from reinfection, Single nucleotide polymorphism (SNP) profiles were compered between the TB episodes. Information on WGS was collected from or (if WGS was not earlier done) performed at the Public Health Agency.
Study definitions
A case of TB recurrence was defined as a patient with a new TB episode, more than 180 days after previously successfully treatment completion. Treatment success was defined as cure if smear- or culture-negative in follow up samples or treatment completion if without evidence of failure, in abundance of follow up samples, according to the WHO definitions (15,16). The anatomical site of TB disease was defined as pulmonary; if only involving the lungs, extrapulmonary; if involving any anatomical site but the lungs (including the pleura) or disseminated; if involving the lungs and any other site (16). The patients country of origin was regarded as endemic at a TB incidence of >100/100,000 population and as low endemic if <100/100,000 (1).
A relapse case was defined as having a maximum difference of five SNPs. This threshold was derived from the proposed cut-off for recent transmission based on earlier studies showing a genetic turnover of 0.3–0.5 SNPs per genome per year (13,17,18).
Resistance to one first line TB-drug, isoniazid (INH), rifampicin, ethambutol or pyrazinamide was regarded as mono drug resistance. Resistance to at least INH and rifampicin was regarded as MDR according to the WHO definitions (15,19).
DNA extraction and WGS
DNA was extracted from M.tb colonies growing on Löwenstein-Jensen medium using a chloroform/CTAB-based protocol or the column-based QIAamp Mini DNA Kit (Qiagen, Hilden, Germany) and the DNA was subsequently quantified with the Qubit dsDNA BR Assay (Thermo Fisher Scientific, Waltham Massachusetts, USA). Sequencing was performed with an Ion Torrent S5 XL instrument (Thermo Fisher Scientific Inc., Waltham, MA, USA) according to the manufacturer’s instructions (read length approximately 300 base pairs) (20,21). The sequences have been deposited in the European Nucleotide Archive under study accession number PRJEB38721.
Single nucleotide polymorphism analysis
The similarity between isolates from the same patient was studied by SNP analysis. Briefly, the generated sequencing reads were mapped against the reference genome H37Rv NC_000962.3 (CLC Assembly Cell version 4.4.2, Qiagen, Hilden, Germany). Variants were extracted (CLC Assembly Cell version 4.4.2), filtered (sequencing depth: ≥10×; frequency: ≥0.9) and converted into a multiple sequence alignment from which a distance matrix was calculated.
Detection of resistance mutations
In order to detect resistance mutations, the sequencing reads were mapped against a set of resistance genes (rpoB, katG, inhA including the promoter region, embB, pncA, gyrA, gyrB, rrs, eis including the promoter region, tlyA and ethA) derived from the reference genome H37Rv NC_000962.3 (CLC Assembly Cell version 4.4.2, Qiagen, Hilden, Germany). The extracted variants (CLC Assembly Cell version 4.4.2, Qiagen, Hilden, Germany) were filtered (minimum frequency of reads calling SNPs: 10%; minimum frequency of reads calling InDels: 80%) and the remaining variants were compared with an in-house database of resistance mutations (based on high confidence and established resistance mutations) (22-24).
Similarly, M.tb lineages were assigned as previously reported (25). A similar analysis was also performed manually in CLC Genomics Workbench 12 version 12.0.3 (Qiagen, Germany).
Statistical analysis
Data was analyzed and plots were created using SPSS (version 25; IBM Corp., Armonk, NY, USA). Continuous data are presented as median and interquartile ranges (IQRs). Comparisons were made by non-parametric tests (Mann-Whitney). Categorical data are presented as frequencies and proportions and comparisons were made by Chi-square tests. Logistic regression analysis was performed to analyze the association of patient characteristics with recurrent TB. The association with the following variables: sex (male, female), age (<35, ≥35), TB-incidence in country of origin (<100/100,000, ≥100/100,000) and clinical manifestation of TB (disseminated, pulmonary, extrapulmonary) was tested in bivariate and multivariate analyses. The associations were presented as odds ratios (OR) with 95% confidence intervals (CI).
To calculate the relapse incidence, the number of relapse cases was divided with the time at risk (follow-up time minus 180 days).
Results
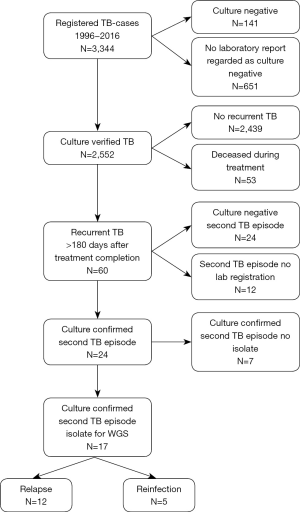
A total of 3,344 TB patients were registered in Stockholm County 1996–2016. Culture verification was obtained in 2,552 cases. Among the 2,552 culture positive cases: 54% were male, 82% were of foreign origin, and 53% were originally from an area with a TB incidence of ≥100/100,000 population. The median age at TB-diagnosis was 35 years (IQR, 26–51 years) (Table 1).
Table 1
| Patients characteristics | Non TB recurrence (n=2,528) | TB recurrence (n=24) | P value | Relapse (n=12) | Reinfection (n=5) |
|---|---|---|---|---|---|
| Median age at TB-diagnosis, years, [IQR] | 35 [26–51] | 24 [19–38] | 0.04 | 27[19–47] | 24 [20–33] |
| Age, years, n [%] | |||||
| <35 | 1,323 [52] | 17 [71] | 0.11 | 8 [67] | 4 [80] |
| ≥35 | 1,205 [48] | 7 [29] | 4 [33] | 1 [20] | |
| Sex, n [%] | |||||
| Female | 1,162 [46] | 12 [50] | 0.85 | 5 [42] | 3 [60] |
| Male | 1,366 [54] | 12 [50] | 7 [58] | 2 [40] | |
| TB-incidence in country of origin, n [%] | |||||
| <100/100,000 | 1,196 [47] | 9 [38] | 0.45 | 5 [42] | 1 [20] |
| ≥100/100,000 | 1332 [53] | 15 [62] | 7 [58] | 4 [80] | |
| Clinical manifestation of TB, n [%] | |||||
| Pulmonary | 1335 [53] | 12 [50] | 0.42 | 6 [50] | 2 [40] |
| Extrapulmonary | 885 [35] | 7 [29] | 3 [25] | 2 [40] | |
| Disseminated | 308 [12] | 5 [21] | 3 [25] | 1 [20] |
TB incidence is expressed in numbers per 100,000 population. Statistically significant values (P value <0.05) are in italic. IQR, interquartile range.
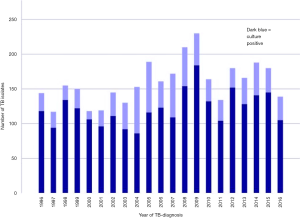
The annual numbers of culture verified cases ranged between a minimum of 85 in 2004 to a maximum of 184 in 2009 (Figure 1). The median follow-up time in the study was 10.2 years (IQR, 5.6–16.2 years). TB reoccurred more than 180 days after successful treatment completion in 60 patients. The second episode was culture confirmed in 24/3,344 cases (0.7%). Out of the 24 cases with a culture confirmed second episode the isolate could be obtained for WGS in 17 patients but in seven patients the second isolate could not be identified at the laboratory. The WGS analysis resulted in 12 cases of relapse and five with reinfection (Figure 2).
In the group of 24 culture verified recurrent cases the median age was 24 (IQR, 19–38) years, 12 (50%) were women, 20 (83%) were born abroad and 15 (62%) were originally from a region classified as a high incidence area. The median age was significantly lower (P=0.04) in the group of patients with TB-recurrence (Table 1).
In a bivariate logistic regression analysis we found no statistically significant associations between TB recurrence and clinical characteristics at TB-diagnosis (Table 2).
Table 2
| Patient characteristics | Non TB recurrence (n=2,528) | TB recurrence (n=24) | Bivariate analysis, OR (95% CI) | P value | Multivariate analysis, OR (95% CI) | P value |
|---|---|---|---|---|---|---|
| Age (years) | ||||||
| <35 | 1,323 | 17 | Reference | |||
| ≥35 | 1,205 | 7 | 2.21 (0.91–5.4) | 0.08* | 2.20 (0.91–5.34) | 0.08 |
| Sex | ||||||
| Female | 1,162 | 12 | Reference | |||
| Male | 1,366 | 12 | 1.18 (0.53–2.63) | 0.52 | 1.16 (0.52–2.59) | 0.72 |
| TB-incidence country of origin | ||||||
| <100/100,000 | 1,196 | 9 | Reference | Not included | ||
| ≥100/100,000 | 1,332 | 15 | 1.57 (0.46–5.38) | 0.47 | ||
| Clinical manifestation of TB | ||||||
| Disseminated | 308 | 5 | Reference | Not included | ||
| Pulmonary | 1,335 | 12 | 2.46 (0.72–8.47) | 0.15 | ||
| Extrapulmonary | 885 | 7 | 2.30 (0.61–8.61) | 0.22 |
*, variables with a P value >0.10 in bivariate analysis were not included in the multivariate analysis except for sex that was considered to be a potential confounding factor. Pulmonary: only lungs involved; Extrapulmonary: lungs not involved; Disseminated: both pulmonary and extrapulmonary involvement. OR, odds ratio; CI, confidence interval.
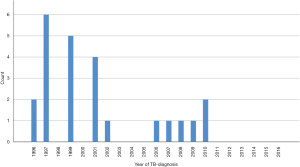
In the recurrent cases (n=24) the median time between the first and the second TB episode, was 3.5 years (IQR, 1.8–5.8 years). In 13/24 (54%) patients, recurrence occurred within four years from the first episode. No case of TB-recurrence was noted in patients diagnosed with their first TB episode 2011–2016 (Figure 3).
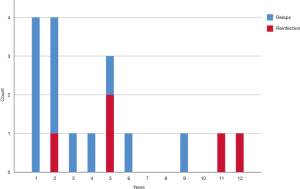
In patients analyzed with WGS (n=17) the median time between treatment completion and the second TB episode was 2.4 years (IQR, 1.1–4.7 years), in relapse cases (n=12) and in reinfected cases (n=5) 5.4 years (IQR, 3.4–11.3 years) (Figure 4).
Results of WGS
The M.tb isolates from the 17 patients with recurrent TB underwent WGS. The Euro-American lineage was the most common lineage, seen in 12/17 (71%) of all recurrent cases and 9/12 (75%) in all relapse cases. The number of SNPs differed from none to 1439 between the first and the second isolate. Although a relapse case was defined as having a maximum difference of five SNPs. For relapse case 1 (Table 3) a difference of seven SNPs was observed but none of these seven SNPs were located in highly variable regions and six of them were not detectable even at very low frequencies in the first isolate (analysis performed to rule out a mixed infection in the first episode). The remaining SNP was more ambiguous and when the SNP analysis was repeated with another reference genome (CP003248.2), the difference between the first and the second isolate was reduced to six. For relapse case 3 the initial SNP analysis revealed a 125 SNP difference. Further analysis did however show a low frequency contamination with a non-mycobacterial species in the rrs gene. When the SNP ratio cut-off was increased to 96%, the contamination was removed and only one SNP remained. Thus, 12 (71%) were classified as relapse and five were regarded as reinfection with a new strain, resulting in a relapse frequency of 0.5% for the whole study period corresponding to an annual risk of 0.06% per year (Table 3).
Table 3
| Case number | Country of origin | Number of single nucleotide poly-morphisms | First episode, drug resistance, gene mutation | Whole genome sequencing lineage, first episode | Second episode, drug resistance, gene mutation | Whole genome sequencing lineage, second episode |
|---|---|---|---|---|---|---|
| Relapse 1 | Sweden | 7 | katG: nt 1-1071 deleted | lineage4.1.2.1 Euro-American | katG: nt 1-1071 deleted | lineage4.1.2.1 Euro-American |
| Relapse 2 | Sweden | 0 | – | lineage4.1.2 Euro-American | – | lineage4.1.2 Euro-American |
| Relapse 3 | Sweden | 1 [125]a | – | lineage4.3.3 Euro-American | – | lineage4.3.3 Euro-American |
| Relapse 4 | Afghanistan | 0 | – | lineage3.1 East-African-Indian | – | lineage3.1 East-African-Indian |
| Relapse 5 | Sweden | 3 | – | lineage4.3.3 Euro-American | – | lineage4.3.3 Euro- |
| Reinfection 1 | Peru | 311 | katG: Ser315Thr; rpoB: Ser450Leu | lineage4.3.3 Euro-American | – | lineage4.3.3 Euro-American |
| Reinfection 2 | Uzbekistan | 155 | – | lineage2.2.1 East-Asian | – | lineage2.2.1 East-Asian |
| Reinfection3 | Kenya | 1317 | – | lineage3.1 East-African-Indian | inhA: C-15T | lineage4.6.1.2 Euro-American |
| Reinfection 4 | Peru | 303 | katG: Ser315Thr; rpoB: Ser450Leu | lineage4.3.3 Euro-American | – | lineage4.3.3 Euro-American |
| Relapse 6 | Somalia | 1 | inhA: C-15T | lineage4.6.1.2 Euro-American | inhA: C-15T | lineage4.6.1.2 Euro-American |
| Relapse 7 | Azerbaijan | 0 | katG: Ser315Thr; rpoB: Ser450Leu; embB:Gly406Asp; pncA: 286-InsT | lineage2.2.1 East-Asian | katG: Ser315Thr; rpoB: Ser450Leu; embB: Gly406Asp; pncA: 286-InsT | lineage2.2.1 East-Asian |
| Relapse 8 | Somalia | 4 | inhA: C-15T | lineage4.6.1.2 Euro-American | inhA: C-15T | lineage4.6.1.2 Euro-American |
| Relapse 9 | Somalia | 1 | inhA: C-15T | lineage4.6.1.2 Euro-American | inhA: C-15T | lineage4.6.1.2 Euro-American |
| Relapse 10 | Somalia | 3 | katG: Ser315Thr; rpoB: Ser450Leu; embB: Met306Val | lineage4 Euro-American | katG: Ser315Thr; rpoB: Ser450Leu; embB: Met306Val | lineage4 Euro-American |
| Relapse 11 | Somalia | 1 | – | lineage2.2.1 East-Asian | – | lineage2.2.1 East-Asian |
| Reinfection 5 | Mongolia | 1,439 | inhA: C-15T | lineage4.3.3 Euro-American | – | lineage2.2.1 East-Asian |
| Relapse 12 | Bolivia | 3 | – | lineage4 Euro-American | – | lineage4 Euro-American |
a, this sample was contaminated with another bacterial species. If the SNP ratio cut-off was increased to 96%, only one SNP remained.
Risk factors for TB recurrence
Drug resistance was present in 9 (53%) of the cases with recurrent TB. In the 12 patients with TB relapse, 4 (33%) had an INH resistant strain and 2 (17%) had an MDR strain. No acquired drug resistance was detected in isolates of the second episode among relapse cases (Table 4).
Table 4
| Characteristic | Relapse (n=12) | Reinfection (n=5) |
|---|---|---|
| Resistance, n [%] | ||
| Mono drug resistance | 4 [33] | 1 [20] |
| Multi drug resistance | 2 [16] | 2 [40] |
| Smoking, n [%] | 6 [50] | 1 [20] |
| Alcohol or drug abuse, n [%] | 3 [25] | 0 |
| HIV infection, n [%] | 0 | 0 |
| Adherence problems, n [%] | 4 [33] | 0 |
Mono drug resistance including resistance to one first line TB-drug. Multidrug resistance including at least resistance to rifampicin and isoniazid.
Among patients with relapse, 4 (36%) had adherence problems, 5 (46%) were smokers and 2 (18%) had alcohol or drug abuse. One patient with several risk factors for TB relapse, relapsed (3 SNP’s difference) after 10 years. In the group of reinfected patients, all were foreign born and 2 (33%) had traveled to their country of origin the year before the second episode. None of the patients with TB recurrence was HIV-co-infected.
Discussion
In this study we showed that only 0.7% of the TB patients diagnosed in Stockholm County between 1996 and 2016 did have a recurrent culture confirmed infection during a median follow-up time more than ten years. Moreover, we found that the majority of the recurrent TB cases, analyzed with WGS (n=17), could be classified as relapse infection with the same strain (n=12) while the remaining patients were considered to be reinfection with a different strain (n=5). When comparing the isolates from the episodes in the 12 relapse cases no acquired drug resistance was detected.
A recent state of the art paper from Rosser concluded that the median percentage of TB patients experiencing a recurrent infection, in studies covering recent decades from low disease burden regions, was 3.4% during a median follow up time of 7.8 years. The somewhat lower recurrence rate in our study is by no means unique and conformingly a recently published national cohort study from Finland covering 1995–2013 demonstrated a similar recurrence rate of 0.6%. Comparison of recurrence rates with earlier studies should though be interpreted with caution as there is no general consensus on the definition of TB recurrence. In our study we regarded recurrent infection as an event occurring more than six months after treatment completion, in consistency with most former studies. In the Finnish study recurrent infection was defined as an infection occurring more than a year after finishing TB treatment. Earlier studies have used different time frames (0–365 days) after treatment completion (17,26-28).
Although most patients in our cohort were diagnosed with a recurrent infection within the first years after TB treatment. In consistency with earlier studies in low endemic settings, we show that relapse can occur several years after treatment completion (3,7). We observed a high frequency of resistant TB strains both among patients with relapse (49%) and reinfection (60%). This is to be compared with the mean frequency of TB resistance of only 13.7% (MDR resistance 2.4%, mono-resistance 11.3%) reported by the Department of Communicable Disease Control and Prevention in Stockholm County during the latter part of our study period 2003–2016 (29). The much higher prevalence of drug resistance among patients with recurrent TB in our cohort indicates that patients infected with drug resistance strains are at larger risk of disease recurrence. Our observation is in consistency with former studies from low endemic settings showing resistant M.tb seems to be a strong risk factor for TB recurrence (7,30-35). Although only a small proportion of all TB patients will develop recurrent infection it is important to acknowledge that it appears at a higher frequency in selected groups and that this might occur several years after treatment completion. Our and earlier studies indicate that patients with resistant TB should be regarded as high risk patients and thus needs improved treatment control and a longer follow-up time for early detection of potential recurrent disease.
M.tb has been reported to have a high genomic stability with a steady mutation rate of around 0.3–0.5 SNPs per genome per year (17,36). The number of differences in SNPs is there for a useful tool to discriminate relapse from reinfection. Although, we consider a five SNP cut-off to define a relapse case, one specific case had isolates differing by 7 SNPs (six when the analysis was repeated with a different reference genome) and was also classified as relapse. This patient was diagnosed with an INH-resistant isolate, treated for 12 months and was then culture positive again 9 months after finalizing treatment. A second relapse episode occurred 12 months after the first relapse. The isolates from the second and the third TB episode did not differ in the WGS analysis (0 SNP). Therefore we conclude that it is important to consider epidemiological and bacteriological information in addition to genotypic data when discriminating between relapse and reinfection. This approach may also apply when outlining clusters and recent transmission.
With earlier used technics for genotyping, reinfection has been considered as the main cause for recurrent infection in high endemic countries (8,9). In contrast relapse has been considered more frequent in low endemic regions (6). As WGS has a higher resolution than earlier used methods, some cases regarded as relapse with former genotyping methods, would be regarded as reinfection with WGS (10). Also, WGS has the capacity to detect intra-host variability and to better identify minority populations that may explain the difficulties in distinguishing relapse from reinfection in patients with a mixed primary infection (this does however call for strict and validated quality criteria) (17,28,37).
There are to our knowledge only two former studies that have used WGS in a low endemic setting to discriminate relapse from reinfection. The formerly mentioned Finnish study and a recent study from New South Wales, Australia both demonstrated that the large majority (80–87%) of disease recurrence was caused by TB relapse (26,27). Although, we also found that relapse was the more common cause for recurrent infection in Stockholm, the proportion of patients that were reinfected in our cohort was somewhat larger than in the Finnish study. This might be an expected finding in a cohort where the majority of TB patients (82%) had their origin from high endemic regions compared to the Finnish cohort where only 14% of the patients were of foreign origin (7). A higher prevalence of TB in some immigrant communities in combination with socioeconomic risk factors as crowded living and more frequent traveling to TB-endemic regions could be expected to increase the risk of reinfection. As many as 33% of the reinfected patients in our cohort had traveled to their country of origin less than one year before their second TB episode.
When interpreting the results from WGS it is important to understand that the analysis does not have the capacity to clearly separate relapse cases reinfection with the same strain. This is illustrated by two patients in our cohort who, according to WGS, were considered relapse cases but who both had partners that were diagnosed with active TB with the same M.tb strain prior to the patients’ relapse. It thus seems plausible that the proportion of reinfection is underestimated when relying only on genotyping methods. These findings underline that prevention of recurrent infection also involves careful and extensive contact tracing.
Our study has several limitations. We could not find culture result for a large proportion of the patients that were registered as TB patients by the handling clinicians. Although, it is possible that the laboratory has failed to register some positive culture results it seems more plausible that these patients rather represent suspected cases under investigation. In our study we have therefore regarded these patients as culture negative which might have underestimated the true recurrence rate. We also could not with accuracy calculate the patient time at risk, as we did not have information, for all included TB-patients, on eventual death or if people moved to another country. Most TB patients are relatively young why only few patients could be expected to die during the study period. Moreover we could not accurately estimate the strength of the association of primary infection with a drug resistant strain with the risk of recurrent infection, as we did only have culture data for the patients with recurrent infection. Furthermore we lacked data on several patient and disease characteristics for the culture positive TB-cohort as a whole why their association with risk of TB relapse could not be calculated. In 24 of the registered 60 cases with a second TB episode, the second case was not culture verified. This could be explained by early diagnosis made on a positive PCR result or clinical suspicion, without culture confirmation. Relapse could not be distinguished from reinfection in this group. PCR can also remain positive several months after treatment completion without viable M.tb.
Conclusions
The frequency of TB recurrence in Stockholm County is low, indicating an overall well-functioning TB-care. WGS seem to be a useful tool to discriminate relapse cases from reinfection cases and could be used to improve care of TB patients and disease control measures. The high proportion of resistant TB strains in patients with relapsing infection indicates a need of better treatment control and longer follow-up time in this group. The substantial proportion of reinfection among patients with relapsing TB in our study underscores that also careful contact tracing is of uttermost importance to limit the risk of reinfection.
Acknowledgments
A draft of this manuscript was included in M Norrbys thesis (Clinical and prophylactic studies of human tuberculosis in a low-endemic setting) published 2019 by Karolinska Institutet.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/jphe-20-10
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jphe-20-10
Peer Review File: Available at http://dx.doi.org/10.21037/jphe-20-10
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jphe-20-10). The authors have no conflicts of interest to declare.
Ethical statement: The authors are accountable for all aspects of work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Ethical approval for the study was obtained from the Regional Ethical Review Board in Stockholm (Dnr 2018/1171-31/1). Informed consent was not obtained as no individuals could be identified through the presented data and as a requirement of informed consent would have introduced uneven response bias in the setting of this population based study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- World Health Organization. Global tuberculosis report 2018. Geneva: World Health Organization, 2018. Licence: CC BY-NC-SA 3.0 IGO.
-
The Public Health Agency of Sweden 2018 . Available online: https://www.folkhalsomyndigheten.se/folkhalsorapportering-statistik/statistikdatabaser-och-visualisering/sjukdomsstatistik/tuberkulos/ - Bang D, Andersen AB, Thomsen VO, et al. Recurrent tuberculosis in Denmark: relapse vs. re-infection. Int J Tuberc Lung Dis 2010;14:447-53. [PubMed]
- Caminero JA, Pena MJ, Campos-Herrero MI, et al. Exogenous reinfection with tuberculosis on a European island with a moderate incidence of disease. Am J Respir Crit Care Med 2001;163:717-20. [Crossref] [PubMed]
- Dobler CC, Crawford AB, Jelfs PJ, et al. Recurrence of tuberculosis in a low-incidence setting. Eur Respir J 2009;33:160-7. [Crossref] [PubMed]
- Rosser A, Marx FM, Pareek M. Recurrent tuberculosis in the pre-elimination era. Int J Tuberc Lung Dis 2018;22:139-50. [Crossref] [PubMed]
- Korhonen V, Soini H, Vasankari T, et al. Recurrent tuberculosis in Finland 1995-2013: a clinical and epidemiological cohort study. Bmc Infect Dis 2017;17:721. [Crossref] [PubMed]
- van Rie A, Warren R, Richardson M, et al. Exogenous reinfection as a cause of recurrent tuberculosis after curative treatment. N Engl J Med 1999;341:1174-9. [Crossref] [PubMed]
- Shen G, Xue Z, Shen X, et al. The study recurrent tuberculosis and exogenous reinfection, Shanghai, China. Emerg Infect Dis 2006;12:1776-8. [Crossref] [PubMed]
- Gardy JL, Johnston JC, Ho Sui SJ, et al. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med 2011;364:730-9. [Crossref] [PubMed]
- Shea J, Halse TA, Lapierre P, et al. Comprehensive Whole-Genome Sequencing and Reporting of Drug Resistance Profiles on Clinical Cases of Mycobacterium tuberculosis in New York State. J Clin Microbiol 2017;55:1871-82. [Crossref] [PubMed]
- Macedo R, Nunes A, Portugal I, et al. Dissecting whole-genome sequencing-based online tools for predicting resistance in Mycobacterium tuberculosis: can we use them for clinical decision guidance? Tuberculosis (Edinb) 2018;110:44-51. [Crossref] [PubMed]
- Walker TM, Kohl TA, Omar SV, et al. Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. Lancet Infect Dis 2015;15:1193-202. [Crossref] [PubMed]
- Gisslén M, Svedhem V, Lindborg L, et al. Sweden, the first country to achieve the Joint United Nations Programme on HIV/AIDS (UNAIDS)/World Health Organization (WHO) 90-90-90 continuum of HIV care targets. HIV Med 2017;18:305-7. [Crossref] [PubMed]
- World Health Organization, Definitions and reporting framework for tuberculosis – 2013 revision 2013 (updated December 2014 and January 2020). Available online: who.int/iris/bitstream/handle/10665/79199/9789241505345_eng.pdf;jsessionid=C722546311D270A8F40680732EB889C8
- World Health Organization. Royal Netherlands Tuberculosis Association. Revised international definitions in tuberculosis control. Int J Tuberc Lung Dis 2001;5:213-5.
- Bryant JM, Harris SR, Parkhill J, et al. Whole-genome sequencing to establish relapse or re-infection with Mycobacterium tuberculosis: a retrospective observational study. Lancet Respir Med 2013;1:786-92. [Crossref] [PubMed]
- Meehan CJ, Moris P, Kohl TA, et al. The relationship between transmission time and clustering methods in Mycobacterium tuberculosis epidemiology. EBioMedicine 2018;37:410-6. [Crossref] [PubMed]
- World Health Organization. TB HIV Collaborative TB/HIV activities 2014. Available online: http://www.who.int/tb/areas-of-work/tb-hiv/tb-hiv_slides_from_2015_global_tb_report.pdf?ua=1
- van Soolingen D, Hermans PW, de Haas PE, et al. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol 1991;29:2578-86. [Crossref] [PubMed]
- Köser CU, Bryant JM, Becq J, et al. Whole-genome sequencing for rapid susceptibility testing of M. tuberculosis. N Engl J Med 2013;369:290-2. [Crossref] [PubMed]
- Miotto P, Tessema B, Tagliani E, et al. A standardised method for interpreting the association between mutations and phenotypic drug resistance in Mycobacterium tuberculosis. Eur Respir J 2017;50:1701354 [Crossref] [PubMed]
- Safi H, Lingaraju S, Amin A, et al. Evolution of high-level ethambutol-resistant tuberculosis through interacting mutations in decaprenylphosphoryl-beta-D-arabinose biosynthetic and utilization pathway genes. Nat Genet 2013;45:1190-7. [Crossref] [PubMed]
- Miotto P, Cabibbe AM, Feuerriegel S, et al. Mycobacterium tuberculosis pyrazinamide resistance determinants: a multicenter study. MBio 2014;5:e01819-14. [Crossref] [PubMed]
- Coll F, McNerney R, Guerra-Assuncao JA, et al. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun 2014;5:4812. [Crossref] [PubMed]
- Korhonen V, Smit PW, Haanpera M, et al. Whole genome analysis of Mycobacterium tuberculosis isolates from recurrent episodes of tuberculosis, Finland, 1995-2013. Clin Microbiol Infect 2016;22:549-54. [Crossref] [PubMed]
- Parvaresh L, Crighton T, Martinez E, et al. Recurrence of tuberculosis in a low-incidence setting: a retrospective cross-sectional study augmented by whole genome sequencing. Bmc Infect Dis 2018;18:265. [Crossref] [PubMed]
- Witney AA, Bateson AL, Jindani A, et al. Use of whole-genome sequencing to distinguish relapse from reinfection in a completed tuberculosis clinical trial. BMC Med 2017;15:71. [Crossref] [PubMed]
- Department of Communicable Disease Control and Prevention SCC. Statistics on Tuberculosis. Available online: https://www.vardgivarguiden.se/behandlingsstod/smittskydd/sjukdomar/statistik/?page=2
- Kim L, Moonan PK, Heilig CM, et al. Factors associated with recurrent tuberculosis more than 12 months after treatment completion. Int J Tuberc Lung Dis 2016;20:49-56. [Crossref] [PubMed]
- Pettit AC, Kaltenbach LA, Maruri F, et al. Chronic lung disease and HIV infection are risk factors for recurrent tuberculosis in a low-incidence setting. Int J Tuberc Lung Dis 2011;15:906-11. [Crossref] [PubMed]
- Selassie AW, Pozsik C, Wilson D, et al. Why pulmonary tuberculosis recurs: a population-based epidemiological study. Ann Epidemiol 2005;15:519-25. [Crossref] [PubMed]
- Millet JP, Orcau A, de Olalla PG, et al. Tuberculosis recurrence and its associated risk factors among successfully treated patients. J Epidemiol Community Health 2009;63:799-804. [Crossref] [PubMed]
- Colangeli R, Jedrey H, Kim S, et al. Bacterial factors that predict relapse after tuberculosis therapy. N Engl J Med 2018;379:823-33. [Crossref] [PubMed]
- El Sahly HM, Wright JA, Soini H, et al. Recurrent tuberculosis in Houston, Texas: a population-based study. Int J Tuberc Lung Dis 2004;8:333-40. [PubMed]
- Guerra-Assunção JA, Houben RM, Crampin AC, et al. Recurrence due to relapse or reinfection with Mycobacterium tuberculosis: a whole-genome sequencing approach in a large, population-based cohort with a high HIV infection prevalence and active follow-up. J Infect Dis 2015;211:1154-63. [Crossref] [PubMed]
- Sobkowiak B, Glynn JR, Houben R, et al. Identifying mixed Mycobacterium tuberculosis infections from whole genome sequence data. BMC Genomics 2018;19:613. [Crossref] [PubMed]
Cite this article as: Norrby M, Groenheit R, Mansjö M, Zedenius I, Vesterbacka J, Lindquist L, Berggren I. Whole genome sequencing of recurrent tuberculosis in Stockholm County 1996–2016. J Public Health Emerg 2020;4:31.