Taking flight with Precision Global Health: a scoping review on avian influenza
Introduction
Avian influenza, also known as ‘bird flu’, is caused by one of the major viruses linking animal populations with humans, with an estimated case fatality rate of 60% (1). Avian influenza viruses consist of segmented negative sense; single stand RNA genomes, derived from the Orthomyxoviridae family (2). These viruses can be further sub-categorized into two groups based on the severity of disease they cause, namely highly pathogenic avian influenza (HPAI) and low pathogenic avian influenza (LPAI) (ibid). Each virus contains one H and one N antigen, and the H5 and H7 strains are known to cause HPAI. It has been hypothesized that HPAI arises from LPAI, due to a faulty polymerase complex resulting in a spontaneous mutation (3). Whilst the latter has been proposed as the most commonly observed pathological pathway, studies have also reported alternative mechanisms, including nucleotide substitutions and recombination with other genes for the emergence of HPAI (ibid).
The natural reservoirs for avian influenza are aquatic birds, which go onto mostly infect domestic poultry and waterfowl, among other bird populations. The virus can be either transmitted through the fecal/oral route, or the respiratory route in land birds (2). Outbreaks of avian influenza are of particular concern in domesticated birds (i.e., poultry) due to the potential to evolve from LPAI to HPAI and death among poultry with HPAI linked to economic losses and trade restrictions (4). In animals, clinical signs for HPAI include sudden onset of high mortalities within flocks associated with edema on the head and face, subcutaneous hemorrhage on the head and wattles and cessation of egg laying (ibid). However, the biggest public health concern is the possibility of the virus to be transmitted to humans. The process in which a disease or infection is transmitted from animal to human, or vice-versa is called zoonosis (1). The first instance of human infection with the avian influenza H5N1 dates back to 1997 in Hong Kong, with 18 identified cases and 6 fatalities, highlighting its pandemic potential (5). Human to human transmission of avian influenza has occurred, with over 200 cases of H7N9 being reported in main land China in 2015 (6). Route of transmission from animal to human is usually via contaminated environments or intermediate hosts, such as pigs, in which exposure may occur through direct contact via slaughtering (6). Conditions for jumping species barriers are ideal in Asia, where poultry, pigs and humans live in crowded conditions, alongside occupational exposure via live poultry markets (3).
In order to mitigate the case of avian influenza, many prevention and control efforts have been put in place, as reducing the risk in animal populations is vital to reduce the risk to humans (1). The so called Tripartite collaboration including WHO, FAO and OIE have set guidelines, and the most common control and prevention measures (7) include: vaccinations of bird populations, legislations such as the OIE Terrestrial Animal Health code (8) and biosecurity measures, which refers to physical and/or procedural measures which may be used to prevent introduction of avian influenza to susceptible poultry (2). The existing prevention and control strategies could be strengthened by the utilization of digital technologies, which may be defined as digital resources which are used to collect novel personal or environmental data (human and animal) from, and by the populations, including but not confined to: mHealth, Big Data and remote-sensing technologies. For instance, spatioal-temporal models may be used to more precisely estimate outbreak distributions, remote-sensing technologies for satellite telemetry, and big data analytics to gain insight into human social and behavioural patterns. Digital technologies thus offer great potential in contributing to control and prevention efforts of avian influenza.
Aims and research question
The aim of this scoping review was to identify the existing literature focused on digital technologies and avian influenza, and to further explore their potential in improving disease monitoring. This scoping review aimed to answer the following question:
What digital technologies were utilized to improve and strengthen detection, control and prevention of avian influenza?
Methodology
A scoping review aims to ‘form knowledge synthesis that addresses an exploratory research question aimed at mapping key concepts, types of evidence, and gaps in research related to a defined area or field by systematically searching, selecting and synthesizing existing knowledge’ (9). This review aimed to map the existing digital technologies used to tackle avian influenza.
Search strategy
The search strategy was developed by three authors, and included a broad range of terms related to digital technologies and avian influenza, which consisted of a combination of free text and MeSH terms (see supplementary appendix). The search terms were used to identify literature that related the use of digital technologies with avian influenza. Disease-related search terms (avian influenza) were identified using MeSH terms from the National Library of Medicine MeSH database, alongside their affiliated catalogued synonyms, whilst digital technology-related search terms were identified through key-terms of a preliminary literature review. Disease-related search terms and digital technology-related search terms were then combined and run in advanced search settings (see Table S1 in supplementary document). For example, a combination of the following search terms but not confined to: [(Avian Influenza) OR (HPAI) OR (H5N1)] AND [(Technology) OR (Big data) OR (Social media) OR (mHealth)], were used to identify relevant literature. Additionally, reference lists of identified material were searched to identify further material of relevance.
Databases
To ensure a comprehensive review of the literature, two databases, namely PubMed and Web of Science were included in the review. Additional literature was identified from grey literature databases utilizing snowball methodology and hand searching previously identified text.
Study selection, inclusion and exclusion criteria
The review considered any studies that discussed the utilization of digital technologies in improving avian-influenza health related outcomes. The review considered peer-reviewed articles (including original quantitative and qualitative studies), but also editorials, viewpoints and letters indexed in PubMed and Web of Science. Text had to be published in English, Spanish, French or German between 2009 (January) and 2017 (July). There were no restrictions with regard to geographic location, population or study design. The review excluded duplicate studies, publication languages other than those specified above, and literature with a strong veterinarian focus opposed to or not linked to public health, with no explicit focus on digital technologies.
Data collection and extraction
Two reviewers independently assessed inclusion and exclusion criteria of titles and abstracts for relevance. The lists of selected literature were then compared between the two reviewers, rationale for inclusion or exclusion was argued, and then selected for the compilation of single list from the two lists previously produced by the two reviewers. Additionally, a third reviewer was involved in the selection process, and also double-checked the final list selected for inclusion. Full text articles were obtained and eligible studies were extrapolated into a descriptive summative table focused on: author, publication date, journal, geographic region, geographic origin of author affiliation, digital technology/device, function, study design, data source, target population, health indicator and challenges. Note that digital technology domains were grouped according to those specified below, created by frequency of emergence (see Table 1). Citations were managed using EndNote software.
Table 1
| Digital technology domain | Description | References |
|---|---|---|
| Big data | A term describing the storage and analysis of large and or complex data sets using a series of techniques including, but not limited to: cloud computing, non-relational databases, natural language processing and machine learning[1] | (10-19) |
| mHealth | Medical and public health practices supported by mobile devices, such as mobile phones, patient monitoring devices, personal digital assistants, and other wireless devices[2] | (20-25) |
| Data modelling | Models involve assumption, abstraction and simplification, of complex disease-associated dynamics[3] | |
| Mathematical modelling | (26-33) | |
| Species and ecological spatial distribution modelling | (34-54) | |
| Suitability and niche modeling | (55-63) | |
| Simulation modelling | (64-77) | |
| Spatioal-temporal modelling | (78-97) | |
| Novel technologies | Case-specific technologies produced or updated, to specifically track and monitor the outbreak, considered “interestingly new or unusual”[4] | (98-112) |
| Remote-sensing technologies | Identifying, observing and measuring an object without coming into direct contact with it[5] | (57,79,113-118) |
[1] Stuart J, Barker A. (2013). Undefined by data: A survey of Big Data definitions. Available online:
Data synthesis
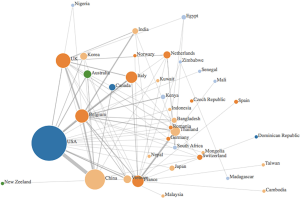
Data was synthesized using a combination of narrative and graphical methods, for a summative description of findings. Additionally, an author’s affiliation network was created to visualize the hubs of digital innovation research in academia. Within the author’s affiliation network, the radius of each circle mirrored the number of publications from each country, the edges were colour based depending on what continent they came from, and the links between countries represented the different collaborations between countries (see Figure 1). The graph was created by adding an edge between the first author and each of the rest of the authors.
Results
Principal findings
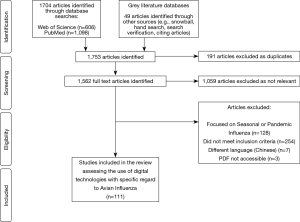
A total of 1,753 titles and abstracts were screened, of which 694 were identified as relevant studies, 191 were excluded as duplicate studies, and 392 did not meet the inclusion criteria. Therefore, a total of 111 studies were selected for inclusion into the review (see Figure 2). Studies included in the review uncovered digital technologies or devices used to tackle avian influenza. Five main themes emerged from the 111 studies in the review, including namely: Big Data, mHealth, data modelling, novel technologies and remote-sensing technologies (see Table 1). Most of the studies were published in 2016, accounting for 21% of publications. Asia persistently had the largest amount of publications (57%), whilst there were no publications included in the review from South America.
The existing research output by country was visualized using an author’s affiliation network (see Figure 1). The highest level of research output was produced by the USA, who was strongly linked with China and Belgium. Other major contributors were based in Europe, namely Belgium, France, Italy and the UK. Many papers cited the Belgium National Fund for Scientific research and the Biological control and Spatial Ecology Unit at the University of Brussels, alongside the FAO, EMPRES wildlife unit for the animal health service based in Italy, which may explain the large contributions of Belgium and Italy, respectively. Many authors countries within the selected articles were affiliated to institutions based in Asian countries—such as Vietnam, India, Korea, Bangladesh, Japan and Cambodia, which was are countries heavily affected by avian influenza.
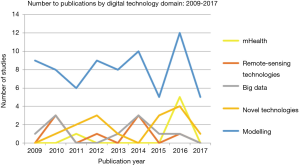
Data modeling
Data modeling accounted for 65% of studies within the review, ranging from computer-assisted mathematical modeling to spatioal-temporal modelling (see Figures 1,3). Mathematical modelling including models based on the Monte-Carlo simulation, Bayesian probabilities, and species distribution models, were mostly used to estimate outbreak distributions, predict host-virus interactions, and more accurately study transmission and control dynamics through various scenarios i.e., live poultry market closures. Additionally, models also yielded a more ecological focus, through species niche modelling, and the use of meteorological data sets to predict and map areas with high probabilities of disease occurrence.
Novel technologies
Novel technologies were also included within the review accounting for 13% of findings, through case-specific diagnostic devices (67%) nanotechnologies (26%) and wearable’s (7%). Novel technologies were mainly utilized for monitoring purposes, with a note-worthy study utilizing nanotechnology for the purpose of treatment.
Big Data
The review identified ten studies (10%) focused on big data, which could be further sub-categorized into social media analytics (40%), web-based surveillance platforms (40%) and online learning resources (20%). Social media platforms were used to capture and inform behavioural changes by measuring user engagement and health communication campaigns. Big data platforms were also utilized for information gathering in the form of web-based surveillance, whilst also supporting online learning.
Remote sensing technologies
A small number of studies explored remote-sensing technologies (7%), under the parameters of satellite telemetry and satellite imagery, capturing how migratory bird populations interacted with their environment, and identifying contaminated bodies of water through earth-satellite observations.
mHealth
A small fraction of research was dedicated to mHealth, constituting 5% of findings within this review. mHealth was mostly used acting as part of a surveillance system, enabled through the SMS or call function for reporting, and GPS technology to track health care workers and cases. However mobile phone devices were also used for diagnostic purposes.
Digital technologies identified within this review were mainly used for surveillance (83%), some dedicated to diagnostics (16%), with low utilization for treatment (1%). Within the surveillance function, data modeling remained dominant, whilst diagnostics were primarily governed by novel technologies and mHealth (see Table 2). Although Big Data was only the third largest domain (9%), it showed great potential in combining multiple data sources.
Table 2
| Digital technology domain | Surveillance (n=92) | Diagnostics (n=18) | Treatment (n=1) |
|---|---|---|---|
| Big data (n=10) | 9 | 1 | 0 |
| mHealth (n=6) | 3 | 3 | 0 |
| Data modelling (n=72) | 71 | 1 | 0 |
| Novel technologies (n=15) | 1 | 13 | 1 |
| Remote-sensing technologies (n=8) | 8 | 0 | 0 |
Discussion
The review identified digital technologies used to tackle avian influenza, with data modelling (65%), under the parameters of computer-assisted mathematical modelling, spatioal-temporal modelling combined with GPS and GIS function being the most utilized. In the case of avian influenza, digital technologies were especially useful in forecasting potential outbreak hotspots by tracking migratory routes and identifying reservoirs through the use of meteorological data sources (55). Most technologies were utilized for surveillance function, with little use of technologies for diagnostic or treatment purposes. Despite avian influenza mainly affecting countries within Asia, high research output was observed in more northern regions, with the exception of China which demonstrated the second highest research output (see Figure 1).
Data modelling was the highest reported utilization of digital technologies within this review (see Table 1). Models were able to predict the most significant variables for disease hotspots, with several studies reporting poultry market density and human population density to be the most significant predictive variables within said model (28,35,64,80). Spatioal-temporal modelling techniques were also combined with global navigation satellite system functions such as GIS and GPS (24%), enabling the identification of route of transmission, and outbreak hotspots through mapping migratory routes of bird populations (31,71). Many of the studies included in data modelling had a strong ecological focus, modelling migratory patterns linked to outbreak occurrence (28,29,38,40,41,57,64,79,80). However, the One Health approach was also incorporated into models, noted by modelling the species jump and assessing risk of human infection (31,57,67,81).
Novel technologies were mainly utilized for the purposes of diagnostics, including diagnostic devices such as the digital microfluidic device, which had the ability to detect a target molecule within tens of seconds (99), RNAi antiviral vector technology (100) and the portable lateral flow device (104). The wearable sensor node was extremely case-specific, as it allowed for poultry to be continuously monitored, alerting administers through the internet when an anomalous state of chickens was detected (110). One noteworthy study was categorized under treatment, seemingly through the use of a novel vaccine using a nanotechnology platform on chickens, which indicated success mirrored in an increased IgG response of the vaccinated chickens when compared to the unvaccinated chickens (98).
Big data was primarily composed of social media analytics, which included text mining of platforms such as Twitter, and also country-specific search engines and blogs referring to Baidu Index and Weibo, respectively (11,15). These social platforms aimed at affecting behavioral change through health communication and increasing user engagement. A prominent theme within the review was the use of data collection through web-based forums, showcasing a participatory approach and collaborative spirit. For instance, the online data platforms CaribVnet and f-FLUA2H both gathered information on avian influenza from both the general population and disease specialists, respectively (12,14). However, a difficulty commonly associated with these forums is the data quality, which may vary by members of the general population. It is important to note that Big Data was also used and generated through online learning tools. For instance, an electronic learning tool specifically tailored for veterinarians focused on avian influenza had a huge success rate, with 90.2% of participants finding online courses useful and convenient, and 97% expected to use the learnt information within their professional lives (18).
Both remote sensing and mHealth represented a small fraction of findings within this review. Remote sensing showcased great potential in capturing migratory patterns and potential hotspots, by utilizing satellite imagery, which identified more contaminated bodies of water which acted as avian influenza reservoirs through earth-satellite observations (113,115,116). Additionally, remote-sensing technologies were able to document poultry market chains through migratory patterns (114). mHealth was mostly utilized for diagnostics purposes, linking mobile phone devices with imaging technologies for a point of use sensing platform (118), but also combining them with fluorescent technologies for a smartphone based fluorescent diagnostic system (21,22).
A major challenge noted in a few of the selected studies (12,79) referenced back to poor biosecurity measures, alongside free-ranging practices which were predominantly highlighted in countries within the Caribbean region and Asia. Within these countries, live poultry markets are a common practice, and as a result guidelines may often not be as closely regulated due to the overarching goal to produce profit. Furthermore, it is also important to consider the economic implications linked to avian influenza, primarily referring to live poultry/bird markets and trade dynamics. As the demand for poultry increases, poultry density and trade activities are also intensified thus increasing the probability of viral spread (119). Embedded within the economic impacts are the underlying anthropogenic factors linked to the more cultural practices, such as the celebration of Luna New year. A recent study found that poultry meat consumption was increased from 4.3- to 9.6-fold during the Luna New year period, exacerbating the cycle of increased demand, increased poultry density and thus increased risk of viral spread (6).
It is important to note that the review also had its methodological limitations, a major one being that only two databases were searched (PubMed and Web of Science), and therefore not capturing the entire evidence body, and also publication bias. Additionally, throughout the eligibility process a large number of studies were excluded due to the focus on seasonal or pandemic influenza, opposed to avian influenza, which may have been caused by the inclusion of “H2N2” within the search strategy syntax (see Table S1 in supplementary documents). The search term was included as some articles; specifically review articles, discussed Influenza as a whole (with the inclusion of avian influenza), and despite the majority of results focusing on pandemic influenza, studies regarding avian influenza and zoonosis were also found and selected for inclusion.
Conclusions
Digital technologies show potential to improve detection, control and prevention for avian influenza. The scoping review mapped the existing digital technologies used to combat avian influenza, and uncovered five main digital domains including: mhealth, Big Data, data modelling, remote-sensing and novel technologies. Results indicated data modelling to be the most utilized technology, primarily used for surveillance purposes. The major hubs of digital innovation, in terms of research output included USA, Belgium and China, presumably due to funding, and high disease prevalence, respectively. It is important to note that although the methodological approach for modeling has advanced by combining computer-assisted simulations with meteorological and remotely sensed data sets, more innovative approaches are still required to fulfill the potential of other existing technologies. It also remains vital to find ways of incorporating these technologies to improve both treatment and diagnostic procedures for avian influenza.
Supplementary
Table S1
| Domain related search terms | Search strategy syntax |
|---|---|
| Digital technology | “Digital” OR “Technology” OR “Precision medicine” OR “Biosensor” OR “Sensors” OR “Bio-surveillance” OR “Intelligent surveillance” OR “Participatory surveillance” OR “Genomic epidemiology” OR “Genomic sequencing” OR “Pathogen genomics” OR “Big data” OR “Data storage” OR “Data science” OR “Information processing” OR “Blockchain” OR “Social media” OR “Twitter” OR “Facebook” OR “Instagram” OR “Flicker” OR “YouTube” OR “Wikipedia” OR “Telemedicine” OR “Robotics” OR “Machine learning” OR “Modelling” OR “Mathematical |
| Avian Influenza | “Influenza in Birds” OR “Influenza, Avian” OR “Fowl Plague” OR “Fowl Plague Virus” OR “Avian Flu” OR “Avian Influenza” OR “Influenza A Virus” OR “Influenza Viruses Type A” OR “Orthomyxovirus Type A” OR “Orthomyxovirus Type A, Avian” OR “Avian Orthomyxovirus Type A” OR “Pestis galli Myxovirus” OR “Myxovirus pestis galli” OR “A (H5N1)” OR “A (H7N9)” OR “A (H9N2)” OR “A (H1N1)” OR “A (H2N2)”OR “Bird Flu” |
Acknowledgments
We would like to acknowledge the Institute of Global Health, Faculty of Medicine at the University of Geneva who supported this work. We would also like to acknowledge and thank Sharada Prasanna Mohanty for his contribution through the production of the authors’ affiliation network.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Mohamed H. Ahmed, Heitham Awadalla and Ahmed O. Almobarak) for the series “The Role of Sudanese Diaspora and NGO in Health System in Sudan” published in Journal of Public Health and Emergency. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jphe.2018.06.01). The series “Precision Infectious Disease Epidemiology” was commissioned by the editorial office without any funding or sponsorship. AF serves as an unpaid editorial board member of Journal of Public Health and Emergency from Apr 2018 to Mar 2020 and served as the unpaid Guest Editor of the series. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- World Health Organization. Avian and other zoonotic influenza 2017. Available online: http://www.who.int/influenza/human_animal_interface/en/, last accessed 07/09/2017.
- Alexander DJ, Capua I. Avian influenza in poultry. World's Poult Sci J 2008;64:513-26. [Crossref]
- Alexander DJ. An overview of the epidemiology of avian influenza. Vaccine 2007;25:5637-44. [Crossref] [PubMed]
- Barman S, Marinova-Petkova A, Hasan MK, et al. Role of domestic ducks in the emergence of a new genotype of highly pathogenic H5N1 avian influenza A viruses in Bangladesh. Emerg Microbes & Infect 2017;6:e72 [Crossref] [PubMed]
- Chan PK. Outbreak of Avian Influenza A(H5N1) Virus Infection in Hong Kong in 1997. Clin Infect Dis 2002;34:S58-64. [Crossref] [PubMed]
- Yang Y, Halloran ME, Sugimoto JD, et al. Detecting human-to-human transmission of avian influenza A (H5N1). Emerg Infect Dis 2007;13:1348-53. [Crossref] [PubMed]
- Food and Agricultural Organization, World Organization for Animal Health and World Health Organization. The Joint FAO–OIE–WHO Global Early Warning System for health threats and emerging risks at the human–animal–ecosystems interface: A concept paper 2013. Available online: http://www.fao.org/3/a-i3579e.pdf, last accessed 30/05/2018
- World Organisation for Animal Health. Terrestrial Animal Health Code 2017. Available online: http://www.oie.int/standard-setting/terrestrial-code/, last accessed 30/05/2018.
- Colquhoun HL, Levac D, O'Brien KK, et al. Scoping reviews: time for clarity, definitions, methods and reporting. J Clin Epidemiol 2014;67:1291-4. [Crossref] [PubMed]
- Kim P, Sorcar P, Um S, et al. Effects of episodic variations in web-based avian influenza education: influence of fear and humor on perception, comprehension, retention and behavior. Health Educ Res 2009;24:369-80. [Crossref] [PubMed]
- Fung IC, Fu KW, Ying Y, et al. Chinese social media reaction to the MERS-CoV and avian influenza A(H7N9) outbreaks. Infect Dis Poverty 2013;2:31. [Crossref] [PubMed]
- Lefrançois T, Hendrikx P, Erhardt N, et al. Surveillance of avian influenza in the Caribbean through the Caribbean Animal Health Network: surveillance tools and epidemiologic studies. Avian Dis 2010;54:369-73. [Crossref] [PubMed]
- Robertson C, Yee L. Avian Influenza Risk Surveillance in North America with Online Media. PLoS One 2016;11:e0165688 [Crossref] [PubMed]
- Sjaugi MF, Tan S, Abd Raman HS, et al. g-FLUA2H: a web-based application to study the dynamics of animal-to-human mutation transmission for influenza viruses. BMC Med Genomics 2015;8:S5. [Crossref] [PubMed]
- Xie T, Yang Z, Yang S, et al. Correlation between reported human infection with avian influenza A H7N9 virus and cyber user awareness: what can we learn from digital epidemiology? Int J Infect Dis 2014;22:1-3. [Crossref] [PubMed]
- Mao C, Wu XY, Fu XH, et al. An internet-based epidemiological investigation of the outbreak of H7N9 Avian influenza A in China since early 2013. J Med Internet Res 2014;16:e221 [Crossref] [PubMed]
- Willeberg P, Perez A, Thurmond M, et al. Visualization and analysis of the Danish 2006 highly pathogenic avian influenza virus H5N1 wild bird surveillance data by a prototype avian influenza BioPortal. Avian Dis 2010;54:433-9. [Crossref] [PubMed]
- Dalla Pozza M, Valerii L, Graziani M, et al. An Electronic Learning Course on Avian Influenza in Italy (2008). Avian Dis 2010;54:784-6. [Crossref] [PubMed]
- Claes F, Kuznetsov D, Liechti R, et al. The EMPRES-i genetic module: a novel tool linking epidemiological outbreak information and genetic characteristics of influenza viruses. Database (Oxford) 2014;2014:bau008 [Crossref] [PubMed]
- Im H, Park YI, Pathania D, et al. Digital diffraction detection of protein markers for avian influenza. Lab Chip 2016;16:1340-5. [Crossref] [PubMed]
- Lin Y, Heffernan C. Accessible and inexpensive tools for global HPAI surveillance: A mobile-phone based system. Prev Vet Med 2011;98:209-14. [Crossref] [PubMed]
- Yeo SJ, Choi K, Cuc BT, et al. Smartphone-Based Fluorescent Diagnostic System for Highly Pathogenic H5N1 Viruses. Theranostics 2016;6:231-42. [Crossref] [PubMed]
- Yeo SJ, Cuc BT, Sung HW, et al. Evaluation of a smartphone-based rapid fluorescent diagnostic system for H9N2 virus in specific-pathogen-free chickens. Arch Virol 2016;161:2249-56. [Crossref] [PubMed]
- Stephenson LM, Biggs JS, Sheppeard V, et al. An evaluation of the use of short message service during an avian influenza outbreak on a poultry farm in Young. Commun Dis Intell Q Rep 2016;40:E195-201. [PubMed]
- Kim HR, Kwon YK, Jang I, et al. Pathologic Changes in Wild Birds Infected with Highly Pathogenic Avian Influenza A(H5N8) Viruses, South Korea, 2014. Emerg Infect Dis 2015;21:775-80. [Crossref] [PubMed]
- Belkhiria J, Alkhamis MA, Martinez-Lopez B. Application of Species Distribution Modeling for Avian Influenza surveillance in the United States considering the North America Migratory Flyways. Sci Rep 2016;6:33161. [Crossref] [PubMed]
- Bui CM, Gardner L, MacIntyer CR, et al. Influenza A H5N1 and H7N9 in China: A spatial risk analysis. PLoS One 2017;12:e0174980 [Crossref] [PubMed]
- Christensen J, El Allaki F, Vallieres A. Adapting a scenario tree model for freedom from disease as surveillance progresses: The Canadian notifiable avian influenza model. Prev Vet Med 2014;114:132-44. [Crossref] [PubMed]
- Dhingra MS, Artois J, Robinson TP, et al. Global mapping of highly pathogenic avian influenza H5N1 and H5Nx clade 2.3.4.4 viruses with spatial cross-validation. Elife 2016;5. [PubMed]
- Gilbert M, Golding N, Zhou H, et al. Predicting the risk of avian influenza A H7N9 infection in live-poultry markets across Asia. Nat Commun 2014;5:4116. [Crossref] [PubMed]
- Li X, Liu X, Xu L, et al. Spatial transmission of avian influenza (type H5) in birds. Integr Zool 2009;4:418-25. [Crossref] [PubMed]
- Shi B, Xia S, Yang GJ, et al. Inferring the potential risks of H7N9 infection by spatiotemporally characterizing bird migration and poultry distribution in eastern China. Infect Dis Poverty 2013;2:8. [Crossref] [PubMed]
- Martcheva M. Avian Flu: Modeling and implications for control. J Bio Sys 2014;22:151-75. [Crossref]
- Alkhamis M, Perez A, Batey N, et al. Modeling the Association of Space, Time, and Host Species with Variation of the HA, NA, and NS Genes of H5N1 Highly Pathogenic Avian Influenza Viruses Isolated from Birds in Romania in 2005-2007. Avian Dis 2013;57:612-21. [Crossref] [PubMed]
- Artois J, Lai S, Feng L, et al. H7N9 and H5N1 avian influenza suitability models for China: accounting for new poultry and live-poultry markets distribution data. Stoch Environ Res Risk Assess 2017;31:393-402. [Crossref] [PubMed]
- Cador C, Rose N, Willem L, et al. Maternally Derived Immunity Extends Swine Influenza A Virus Persistence within Farrow-to-Finish Pig Farms: Insights from a Stochastic Event-Driven Metapopulation Model. PLoS One 2016;11:e0163672 [Crossref] [PubMed]
- Cappelle J, Girard O, Fofana B, et al. Ecological modeling of the spatial distribution of wild waterbirds to identify the main areas where avian influenza viruses are circulating in the Inner Niger Delta, Mali. Ecohealth 2010;7:283-93. [Crossref] [PubMed]
- Chong NS, Tchuenche JM, Smith RJ. A mathematical model of avian influenza with half-saturated incidence. Theory Biosci 2014;133:23-38. [Crossref] [PubMed]
- Farnsworth ML, Fitchett S, Hidayat MM, et al. Metapopulation dynamics and determinants of H5N1 highly pathogenic avian influenza outbreaks in Indonesian poultry. Prev Vet Med 2011;102:206-17. [Crossref] [PubMed]
- Hernandez-Jover. Evaluating the risk of avian influenza introduction and spread among poultry exhibition flocks in Australia. Prev Vet Med 2015;118:128-41. [Crossref] [PubMed]
- Hill EM, House T, Dhingra MS, et al. Modelling H5N1 in Bangladesh across spatial scales: Model complexity and zoonotic transmission risk. Epidemics 2017;20:37-55. [Crossref] [PubMed]
- Lu J, Liu W, Xia R, et al. Effects of closing and reopening live poultry markets on the epidemic of human infection with avian influenza A virus. J Biomed Res 2016;30:112-9. [PubMed]
- Moriguchi S, Onuma M, Goka K. Potential risk map for avian influenza A virus invading Japan. Diversity and Distributions 2013;19:78-85. [Crossref]
- Nickbakhsh S, Hall M, Dorigatti I, et al. Modelling the impact of co-circulating low pathogenic avian influenza viruses on epidemics of highly pathogenic avian influenza in poultry. Epidemics 2016;17:27-34. [Crossref] [PubMed]
- Paul M, Tarvonparnich S, Abrial D, et al. Anthropogenic factors and the risk of highly pathogenic avian influenza H5N1: prospects from a spatial-based model. Vet Res 2010;41:28. [Crossref] [PubMed]
- Pelletier STK, Rorres S, Macko PC, et al. Models of highly pathogenic avian influenza epidemics in commercial poultry flocks in Nigeria and Ghana. Trop Anim Health Prod 2012;44:1681-7. [Crossref] [PubMed]
- Reynolds JJH, Torremorell M, Craft ME. Mathematical Modeling of Influenza A Virus Dynamics within Swine Farms and the Effects of Vaccination. PLoS One 2014;9:e106177 [Crossref] [PubMed]
- Stevens KB, Gilbert M, Pfeiffer DU. Modeling habitat suitability for occurrence of highly pathogenic avian influenza virus H5N1 in domestic poultry in Asia: a spatial multicriteria decision analysis approach. Spat Spatiotemporal Epidemiol 2013;4:1-14. [Crossref] [PubMed]
- Takekawa JY, Hill NJ, Schultz AK, et al. Rapid diagnosis of avian influenza virus in wild birds: use of a portable rRT-PCR and freeze-dried reagents in the field. J Vis Ex 2011;(54).
- Van Boeckel TP, Thanapngtharm W, Robinson T, et al. Improving risk models for avian influenza: the role of intensive poultry farming and flooded land during the 2004 Thailand epidemic. PLoS One 2012;7:e49528 [Crossref] [PubMed]
- Vittecoq M, Gaudin H, Oudart T, et al. Modeling the spread of avian influenza viruses in aquatic reservoirs: A novel hydrodynamic approach applied to the Rhone delta (southern France). Sci Total Environ 2017;595:787-800. [Crossref] [PubMed]
- Wiratsudakul A, Paul MC, Bicout DJ, et al. Modeling the dynamics of backyard chicken flows in traditional trade networks in Thailand: implications for surveillance and control of avian influenza. Trop Anim Health Prod 2014;46:845-53. [Crossref] [PubMed]
- Goutard FL, Paul M, Tavornpanich S, et al. Optimizing early detection of avian influenza H5N1 in backyard and free-range poultry production systems in Thailand. Prev Vet Med 2012;105:223-34. [Crossref] [PubMed]
- Yu H, Wu JT, Cowling BJ, et al. Effect of closure of live poultry markets on poultry-to-person transmission of avian influenza A H7N9 virus: an ecological study. Lancet 2014;383:541-8. [Crossref] [PubMed]
- Biswas PK, Islam Z, Debnth NC, et al. Modeling and roles of meteorological factors in outbreaks of highly pathogenic avian influenza H5N1. PLoS One 2014;9:e98471 [Crossref] [PubMed]
- Boni MF, Manh BH, Thai Q, et al. Modelling the progression of pandemic influenza A (H1N1) in Vietnam and the opportunities for reassortment with other influenza viruses. BMC Med 2009;7:43. [Crossref] [PubMed]
- Dhingra MS, Dissanayake R, Negi AB, et al. Spatio-temporal epidemiology of highly pathogenic avian influenza (subtype H5N1) in poultry in eastern India. Spat Spatiotemporal Epidemiol 2014;11:45-57. [Crossref] [PubMed]
- Hénaux V, Samuel MD, Bunck CM. Model-Based Evaluation of Highly and Low Pathogenic Avian Influenza Dynamics in Wild Birds. PLoS One 2010;5:e10997 [Crossref] [PubMed]
- Loth L, Gilbert M, Osmani MG, et al. Risk factors and clusters of Highly Pathogenic Avian Influenza H5N1 outbreaks in Bangladesh. Prev Vet Med 2010;96:104-13. [Crossref] [PubMed]
- Nishiguchi A, Kobyashi S, Ouchi Y, et al. Spatial Analysis of Low Pathogenic H5N2 Avian Influenza Outbreaks in Japan in 2005. J Vet Med Sci 2009;71:979-82. [Crossref] [PubMed]
- Prosser DJ, Hungerford LL. Spatial Modeling of Wild Bird Risk Factors for Highly Pathogenic A(H5N1) Avian Influenza Virus Transmission. Avian Dis 2016;60:329-36. [Crossref] [PubMed]
- Rao DM, Chernyakhovsky A, Rao V. Modeling and analysis of global epidemiology of avian influenza. Environmental Modelling & Software 2009;24:24-134. [Crossref]
- Takekawa JY, Prosser DJ, Collins BM, et al. Movements of wild ruddy shelducks in the Central Asian Flyway and their spatial relationship to outbreaks of highly pathogenic avian influenza H5N1. Viruses 2013;5:2129-52. [Crossref] [PubMed]
- Ekong PS, Ducheyne E, Carpenter TE, et al. Spatio-temporal epidemiology of highly pathogenic avian influenza (H5N1) outbreaks in Nigeria, 2006-2008. Prev Vet Med 2012;103:170-7. [Crossref] [PubMed]
- Farnsworth ML, Ward MP. Identifying spatio-temporal patterns of transboundary disease spread: examples using avian influenza H5N1 outbreaks. Vet Res 2009;40:20. [Crossref] [PubMed]
- Hagenaars TJ, Fischer EA, Jansen JM, et al. Modelling the Innate Immune Response against Avian Influenza Virus in Chicken. PLoS One 2016;11:e0157816 [Crossref] [PubMed]
- Herrick KA, Huettmann F, Lindgren MA. A global model of avian influenza prediction in wild birds: the importance of northern regions. Vet Res 2013;44:42. [Crossref] [PubMed]
- Kitajima M, Huang Y, Watanabe T, et al. Dose-response time modelling for highly pathogenic avian influenza A (H5N1) virus infection. Lett Appl Microbiol 2011;53:438-44. [Crossref] [PubMed]
- Liang L, Xu B, Chen Y, et al. Combining spatial-temporal and phylogenetic analysis approaches for improved understanding on global H5N1 transmission. PLoS One 2010;5:e13575 [Crossref] [PubMed]
- Marquetoux N, Paul M, Wognarkpet S, et al. Estimating spatial and temporal variations of the reproduction number for highly pathogenic avian influenza H5N1 epidemic in Thailand. Prev Vet Med 2012;106:143-51. [Crossref] [PubMed]
- Prosser DJ, Hungerford LL, Erwin RM, et al. Mapping avian influenza transmission risk at the interface of domestic poultry and wild birds. Front Public Health 2013;1:28. [Crossref] [PubMed]
- Rorres C, Pelletier ST, Bruhn MC, et al. Ongoing estimation of the epidemic parameters of a stochastic, spatial, discrete-time model for a 1983-84 avian influenza epidemic. Avian Dis 2011;55:35-42. [Crossref] [PubMed]
- Van Boeckel TP, Thanapngtharm W, Robinson T, et al. Predicting the distribution of intensive poultry farming in Thailand. Agric Ecosyst Environ 2012;149:144-53. [Crossref] [PubMed]
- Zhang Y, Shen Z, Ma C, et al. Cluster of human infections with avian influenza A (H7N9) cases: a temporal and spatial analysis. Int J Environ Res Public Health 2015;12:816-28. [Crossref] [PubMed]
- Si Y, Skidmore AK, Wang T, et al. Spatio-temporal dynamics of global H5N1 outbreaks match bird migration patterns. Geospat Health 2009;4:65-78. [Crossref] [PubMed]
- Minh PQ, Morris RS, Schauer B, et al. Spatio-temporal epidemiology of highly pathogenic avian influenza outbreaks in the two deltas of Vietnam during 2003-2007. Prev Vet Med 2009;89:16-24. [Crossref] [PubMed]
- Si Y, Skidmore AK, Wang T, et al. Spatio-temporal dynamics of global H5N1 outbreaks match bird migration patterns. Geospat Health 2009;4:65-78. [Crossref] [PubMed]
- Bodbyl-Roels S, Peterson AT, Xiao X. Comparative analysis of remotely-sensed data products via ecological niche modeling of avian influenza case occurrences in Middle Eastern poultry. Int J Health Geogr 2011;10:21. [Crossref] [PubMed]
- Dorjee S, Revie CW, Poljak Z, et al. One-Health Simulation Modelling: Assessment of Control Strategies Against the Spread of Influenza between Swine and Human Populations Using NAADSM. Transbound Emerg Dis 2016;63:e229-e244. [Crossref] [PubMed]
- Fang LQ, Li XL, Liu K, et al. Mapping spread and risk of avian influenza A (H7N9) in China. Sci Rep 2013;3:2722. [Crossref] [PubMed]
- Fasina FO, Njage PM, Ali AM, et al. Development of Disease-specific, Context-specific Surveillance Models: Avian Influenza (H5N1)-Related Risks and Behaviours in African Countries. Zoonoses and Public Health 2016;63:20-33. [Crossref] [PubMed]
- Hill AA, Dewè T, Kosmider R, et al. Modelling the species jump: towards assessing the risk of human infection from novel avian influenzas. R Soc Open Sci 2015;2:150173 [Crossref] [PubMed]
- Jewell CP, Kypraios T, Christley RM, et al. A novel approach to real-time risk prediction for emerging infectious diseases: a case study in Avian Influenza H5N1. Prev Vet Med 2009;91:19-28. [Crossref] [PubMed]
- Li XL, Yang Y, Sun Y, et al. Risk Distribution of Human Infections with Avian Influenza H7N9 and H5N1 virus in China. Sci Rep 2015;5:18610. [Crossref] [PubMed]
- Liu S, Ruan S, Zhang X. Nonlinear dynamics of avian influenza epidemic models. Mathematical Biosciences 2017;283:118-35. [Crossref] [PubMed]
- Martin V, Pfeiffer D, Zhou X, et al. Spatial distribution and risk factors of highly pathogenic avian influenza (HPAI) H5N1 in China. PLoS Pathog 2011;7:e1001308 [Crossref] [PubMed]
- Ojimelukwe AE, Prakarnkamanant A, Rushton J. Estimating the sensitivity of passive surveillance for HPAI H5N1 in Bayelsa state, Nigeria. Prev Vet Med 2016;129:58-66. [Crossref] [PubMed]
- Paul MC, Gourtard FL, Rolleau F, et al. Quantitative assessment of a spatial multicriteria model for highly pathogenic avian influenza H5N1 in Thailand, and application in Cambodia. Sci Rep 2016;6:31096. [Crossref] [PubMed]
- Prosser DJ, Palm EC, Takekawa JY, et al. Movement analysis of free-grazing domestic ducks in Poyang Lake, China: a disease connection. Int J Geogr Inf Sci 2016;30:869-80. [Crossref] [PubMed]
- Ssematimba A, Hagenaars TJ, de Jong MC. Modelling the wind-borne spread of highly pathogenic avian influenza virus between farms. PLoS One 2012;7:e31114 [Crossref] [PubMed]
- Tian H, Dong L, Zhou S, et al. Spatial, temporal and genetic dynamics of highly pathogenic avian influenza A (H5N1) virus in China. BMC Infect Dis 2015;15:54. [Crossref] [PubMed]
- Van Boeckel TP, Prosser D, Fanceschini G, et al. Modelling the distribution of domestic ducks in Monsoon Asia. Agric Ecosyst Environ 2011;141:373-80. [Crossref] [PubMed]
- Zhang Z, Chen D, Ward MP, et al. Transmissibility of the highly pathogenic avian influenza virus, subtype H5N1 in domestic poultry: a spatio-temporal estimation at the global scale. Geospat Health 2012;7:135-43. [Crossref] [PubMed]
- Bentley RA, Ormerod P. A rapid method for assessing social versus independent interest in health issues: a case study of 'bird flu' and 'swine flu'. Soc Sci Med 2010;71:482-5. [Crossref] [PubMed]
- Loth L, Gilbertb M, Mozaffar MG, et al. Risk factors and clusters of Highly Pathogenic Avian Influenza H5N1 outbreaks in Bangladesh. Prev Vet Med 2010;96:104-13. [Crossref] [PubMed]
- Ward MP, Maftei D, Apostu C, et al. Environmental and anthropogenic risk factors for highly pathogenic avian influenza subtype H5N1 outbreaks in Romania, 2005--2006. Vet Res Commun 2008;32:627-34. [Crossref] [PubMed]
- Kim KI, Lin Z, Zhang L. Avian-human influenza epidemic model with diffusion. Nonlinesuiar Analysis: Real World Applications 2010;11:313-22. [Crossref]
- Babapoor S, Neef T, Mittleholzer C, et al. A Novel Vaccine Using Nanoparticle Platform to Present Immunogenic M2e against Avian Influenza Infection. Influenza Res Treat 2011;126794 [PubMed]
- Choi K, Kim JY, Ahn JH, et al. Integration of field effect transistor-based biosensors with a digital microfluidic device for a lab-on-a-chip application. Lab Chip 2012;12:1533-9. [Crossref] [PubMed]
- Huang J, Xie Z, Xie Z, et al. Silver nanoparticles coated graphene electrochemical sensor for the ultrasensitive analysis of avian influenza virus H7. Anal Chim Acta 2016;913:121-7. [Crossref] [PubMed]
- Karash S, Wang R, Kelso L, et al. Rapid detection of avian influenza virus H5N1 in chicken tracheal samples using an impedance aptasensor with gold nanoparticles for signal amplification. J Virol Methods 2016;236:147-56. [Crossref] [PubMed]
- Linke LM, Wilusz J, Pabilonia KL, et al. Inhibiting avian influenza virus shedding using a novel RNAi antiviral vector technology: proof of concept in an avian cell model. Amb Express 2016;6:16. [Crossref] [PubMed]
- Nagy A, Cerníková L, Křivda V, et al. Digital genotyping of avian influenza viruses of H7 subtype detected in central Europe in 2007-2011. Virus Res 2012;165:126-33. [Crossref] [PubMed]
- Soliman M, Selimi M, Coward VJ, et al. Evaluation of two commercial lateral flow devices (LFDs) used for flockside testing of H5N1 highly-pathogenic avian influenza infections in backyard gallinaceous poultry in Egypt. J Mol Genet Med 2010;4:247-51. [Crossref] [PubMed]
- Takekawa JY, Hill, NJ, Schultz AK, et al. Rapid diagnosis of avian influenza virus in wild birds: use of a portable rRT-PCR and freeze-dried reagents in the field. J Vis Exp 2011;(54).
- Vidic J, Manzano M, Chang CM, et al. Advanced biosensors for detection of pathogens related to livestock and poultry. Vet Res 2017;48:11. [Crossref] [PubMed]
- Yan Y, Jia XY, Wang HH, et al. Dynamic quantification of avian influenza H7N9(A) virus in a human infection during clinical treatment using droplet digital PCR. J Virol Methods 2016;234:22-7. [Crossref] [PubMed]
- Ahn JH, Im M, Park TJ, et al. Label-Free and Real-Time Detection of Avian Influenza Using Nanowire Field Effect Transistors. J Biomed Nanotechnol 2015;11:1640-3. [Crossref] [PubMed]
- Lum J, Wang R, Hagris B, et al. An Impedance Aptasensor with Microfluidic Chips for Specific Detection of H5N1 Avian Influenza Virus. Sensors (Basel) 2015;15:18565-78. [Crossref] [PubMed]
- Okada H, Nogami H, Kobyashi T, et al. Avian Influenza Surveillance System with Wearable Wireless Sensor Node Using Pb(Zr, Ti)O-3 Microcantilever. Sensors and Materials 2013;25:619-26.
- Zou X, Huang H, Gao Y, et al. Detection of avian influenza virus based on magnetic silica nanoparticles resonance light scattering system. Analyst 2012;137:648-53. [Crossref] [PubMed]
- Abd El Wahed A, Weidmann M, Hufert FT. Diagnostics-in-a-Suitcase: Development of a portable and rapid assay for the detection of the emerging avian influenza A (H7N9) virus. J Clin Virol 2015;69:16-21. [Crossref] [PubMed]
- Bridge ES, Kelly FS, Xiao X, et al. Bird Migration and Avian Influenza: A Comparison of Hydrogen Stabl Isotopes and Satellite Tracking Methods. Ecol Indic 2014;45:266-73. [Crossref] [PubMed]
- Choi CY, Takekawa J, Xiong Y, et al. Tracking domestic ducks: A novel approach for documenting poultry market chains in the context of avian influenza transmission. J Int Agri 2016;15:1584-94. [Crossref]
- Gaidet N, Cappelle J, Takekawa JY, et al. Potential spread of highly pathogenic avian influenza H5N1 by wildfowl: dispersal ranges and rates determined from large-scale satellite telemetry. J App Ecolo 2010;47:1147-57. [Crossref]
- Gilbert M, Newman S, Takekawa JY, et al. Flying over an infected landscape: distribution of highly pathogenic avian influenza H5N1 risk in South Asia and satellite tracking of wild waterfowl. Ecohealth 2010;7:448-58. [Crossref] [PubMed]
- Guerrini L, Paul MC, Leger L, et al. Landscape attributes driving avian influenza virus circulation in the Lake Alaotra region of Madagascar. Geospat Health 2014;8:445-53. [Crossref] [PubMed]
- Tran A, Goutard F, Chamaillé L, et al. Remote sensing and avian influenza: A review of image processing methods for extracting key variables affecting avian influenza virus survival in water from Earth Observation satellites. International Journal of Applied Earth Observation and Geoinformation 2010;12:1-8. [Crossref]
- Delabouglise A, Choisy M, Phan TD, et al. Economic factors influencing zoonotic disease dynamics: demand for poultry meat and seasonal transmission of avian influenza in Vietnam. Sci Rep 2017;7:5905. [Crossref] [PubMed]
Cite this article as: Bempong NE, Ruiz De Castañeda R, Dietrich D, Bolon I, Flahault A. Taking flight with Precision Global Health: a scoping review on avian influenza. J Public Health Emerg 2018;2:21.