
MicroRNA profiling in plasma of HIV-1 infected patients: potential markers of infection and immune status
Introduction
Over the past decade, acquired immunodeficiency syndrome (AIDS) caused by HIV-1 infection began to spread from the original drug use, commercial sex workers and other high-risk groups to the general population. By the end of 2014, nearly 501,000 HIV/AIDS people (including 59.1% HIV people and 40.9% AIDS patients) and 159,000 deaths had been reported in China (1). However, HIV-1 is continuing to spread, with an estimated rate of about 50,000 new infections annually (1). Rapid and accurate diagnostic tools were essential for early detection and monitoring the immune status of HIV-1 infection. Current laboratory tests can detect HIV-1 antibodies, viral nucleic acids, CD4+ T lymphocytes, and p24 antigens (2). HIV-1 antibody test is the gold standard, but sometimes the false negative results could be detected during the window period of infection. As the main target cells of HIV, the CD4+ T cell counts are useful to monitor how effective antiretroviral treatment (ART) is in suppressing the virus and determine the risk of progression of HIV disease.
Recently, several miRNAs are being used as biomarkers for the diagnosis of infectious diseases. MicroRNAs (miRNAs) are small (21–22 nt), non-coding RNAs which could play important roles in infectious diseases (3,4). Previous studies have found the different expression pattern of miRNAs in PBMCs, CD8+ T cells, monocytes and CD4+ T cells from HIV-1 infected subjects (5-9). Huang et al. has established that five miRNAs including miR-28 and miR-223 can directly target to the 3’ end of HIV mRNAs in order to inhibit viral mRNA expression in CD4+ T cells (5). Wang et al. has reported that miR-223 and miR-28 showed different expression level between monocytes and macrophages in HIV infection (6). Other studies have shown that miR-29a in the PBMC and CD4+ T cells can reduce viral replication (10). Furthermore, circulating miRNAs have been intensively studied in various cancers and infectious diseases (11-16). Serum/plasma miRNAs are stable, resistant to RNase digestion and consistent in the same species which could act as useful biomarkers for disease detection (17,18).
The goal of our study was set to identify differentially expressed miRNAs isolated from HIV-1 individuals that could be used to assist in HIV-1 detection and analyze the potential biological functions of these miRNAs. To further illustrate the correlation between miRNAs expression and HIV/AIDS immune progression, we have separated the HIV-1 infected individuals into three groups according to the CD4+ T cell level and have analyzed the expression level of the candidate miRNAs in these three groups.
Methods
Sample collection
A total of 165 participants (37 healthy subjects and 128 patients with HIV-1 infection) were enrolled in the Jiangsu Province in 2013. The 128 infected people were categorized into low (CD4 <200 cell/ µL) (n=41), medium (200< CD4 <350 cell/µL) (n=44) and high (CD4 >350 cell/ µL) (n=43) groups based on CD4+ T cell count. Among them, ten subjects (sex and age matched) from each group were used in the Taqman Low-Density Array (TLDA) study. All subjects taken by the RT-qPCR test were used for confirming the array data. Blood samples were obtained by venipuncture into BD Vacutainer tubes with EDTA-k2. The samples were centrifuged immediately at 2,000 ×g for 15 minutes at room temperature (RT). Aliquots of plasma were stored in −80 °C until use. Plasma samples were collected from confirmed HIV-1 infected patients without ART who had HIV-1 antibody positive results using HIV Western Blot Assay (MP Biomedicals Asia Pacific Pte Ltd, Singapore) at the time of enrollment. All people were not infected with hepatitis B and C viruses
Analysis of the plasma miRNA profile by TLDA
The plasma pools were created by combining ten samples (30 µL per sample) from four respective groups (control group; three HIV-1 infection groups with low, medium and high CD4+ T cells) for TLDA analysis. Total RNA was isolated from each pool of plasma samples using NucleoSpin miRNA Plasma kits (Macherey-Nagel GmbH & Co, Germany) following the manufacturer’s instructions. miRNA expression profiles were executed by the TLDA v3.0 (Applied Biosystems, USA) which can detect 754 miRNAs including 4 endogenous controls. The TLDA experiment was performed and analyzed as previously reported (19).
Confirmation Candidate miRNAs using RT-qPCR
Total RNA used for RT-qPCR assay was extracted from individual plasma sample following the instruction as above. Cel-miR-238 used as an internal control was added into each individual sample before starting the isolation procedure. To confirm the array results, candidate miRNAs were quantified for each individual plasma sample by RT-qPCR test. The RT-qPCR experiment was performed and analyzed as previously reported (19).
Statistical analysis
Log2 relative level was used to compare the difference between the target miRNA and cel-miR-238 (∆Cq). Student’s t-test was used to compare the differences in the miRNA expression between the two groups. 1-way ANOVA was used to compare between more than two groups and the differences between the groups were performed by the Fisher LSD test. A p value <0.05 shows the statistically significant. In addition, the area under curve (AUC) value and a receiver operating characteristic (ROC) curve were used to evaluate the diagnostic potential of each miRNA. 95% confidence intervals (CI) were used to detect the sensitivity and specificity of HIV-1 infection. In order to increase the accuracy of diagnostics, multiple logistic regression analysis was performed as previously reported (20).
Results and Discussion
Demographic characteristics of HIV-1 infected patient
Table 1 showed the demographic characteristics of HIV-1 infected patients. A total of 165 subjects were taken part in this study including 128 HIV-1 infected patients (87 males and 41 females; median age, 42.70±14.34 years) and 37 healthy people (18 males and 19 females; median age, 39.92±11.59 years). There was a significant difference (P<0.05, chi-square test) on the gender distribution between the HIV-1 infection and healthy people, but there was no significant difference in age between the two groups (P>0.05, t-test). The HIV-1 infected people were categorized into low (CD4 <200 cell/ µL), medium (200< CD4 <350 cell/ µL) and high (CD4 >350 cell/ µL) groups based on CD4+ T cell count. The first two groups showed significant differences (P<0.01 and P<0.05, chi-square test) in gender distribution between the HIV-1 infection and healthy people. There was no significant difference (P>0.05, chi-square test) in gender distribution in high CD4+ T cell count group. Although we found gender differences between the HIV-1 infection and healthy people, we and other study have shown that gender differences have no effect on miRNAs expression (15-17).
Table 1
| Sample characteristic | Patients group | Healthy controls group | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CD4+ T cell counts (<200) | CD4+ T cell counts (200–350) | CD4+ T cell counts (>350) | |||||||||
| TLDA study | Validation study | TLDA study | Validation study | TLDA study | Validation study | TLDA study | Validation study | ||||
| Number of participants | 10 | 41 | 10 | 44 | 10 | 43 | 10 | 37 | |||
| Gender (male/female) | 5/5 | 36/5 | 5/5 | 32/12 | 5/5 | 19/24 | 5/5 | 18/19 | |||
| Age (years, mean ± SD) | 33.50±8.71 | 42.54±16.93 | 34.30±8.76 | 41.52±12.72 | 34.90±8.86 | 44.05±13.42 | 34.50±10.11 | 39.92±11.59 | |||
| CD4+ T cell count (cell/ìL) (mean ± SD) | 99.41±64.88 | 275.09±40.77 | 513.05±154.49 | NA | |||||||
miRNA profiling analysis in control and HIV-1 Infected groups
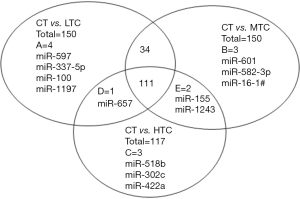
TLDA analysis was executed to identify the differentially expressed level in HIV-1 subjects with low, medium, high CD4+ T cell count (LTC, MTC, and HTC), and control subjects. Results from LTC, MTC, and HTC groups were compared with the control group. Of the 754 host miRNAs (including endogenous controls) incorporated in the array, 231, 345, 315, and 257 miRNAs (Cq values <40) were detected in plasma of healthy controls, LTC, MTC, and HTC groups, respectively. In order to screen out HIV-specific candidate miRNAs, we set up two criteria: (I) Cq values <35 in both of two groups; and (II) Fold change ≥2 between the two groups. Total of 150 miRNAs met the two criteria in LTC subjects, of which 147 were up-regulated and 3 were down-regulated compared to controls (Table S1). Similarly, in the MTC group, a total of 150 miRNAs (148 up-regulated and 2 down-regulated) were differentially regulated compared to healthy controls (Table S2). However, in the HTC group, only 117 miRNAs (115 up-regulated and 2 down-regulated) were differentially regulated compared to healthy controls (Table S3). Figure 1 displays the number and overlap of significantly dysregulated miRNAs which are specific to each group. A total of 112 miRNAs (LTC and HTC combined) showed different expression in HIV-1 infection compared with healthy controls (CT). Similarly, 145 miRNAs (LTC and MTC combined) and 113 miRNAs (MTC and HTC combined) were significantly dysregulated in the infection. Among the 150 miRNAs that were dysregulated between the controls and LTC groups, 4 were unique to the LTC group. When the control was compared with MTC and HTC separately, 3 of the 150 and 3 of the 117 miRNAs were specific to the MTC and HTC groups. While comparing all the three groups, there were 111 different expressed miRNAs in all of them. Based on the HIV infection literatures and the results of target gene analysis, 12 miRNAs (miR-29a, miR-223, miR-27a, miR-19b, miR-766, miR-28-5p, miR-151-3p, miR-30a-3p, miR125b, miR-18a, miR-1197 and miR-518b) were selected for further analysis. Among these, 9 miRNAs (miR-223, miR-19b, miR-27a, miR-30a-3p, miR-151-3p, miR-766, miR-28-5p, miR-125b, and miR-18a) were commonly up-regulated in all three groups. Mir-29a was up-regulated expression in LTC and MTC groups but not in HTC group. MiR-1197 and miR-518b were unique in LTC and HTC groups, separately.
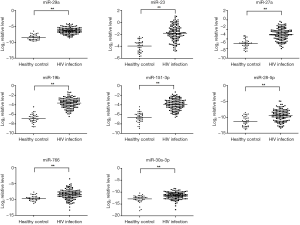
RT-qPCR confirmation of miRNA expression in HIV-1 infected subjects
RT-qPCR (TaqMan miRNA assays) was used to confirm the expression levels of 12 candidate miRNAs which were identified by TLDA. Eight miRNAs (miR-29a, miR-19b, miR-223, miR-27a, miR-151-3p, miR-766, miR-28-5p, and miR-30a-3p) were significant up-regulation in HIV-1 infected plasma (P<0.05, student’s t-test) (Figure 2) compared to healthy control. However, there was no significant difference in other four miRNAs (miR-1197, miR-125b, miR18a, and miR-518b) in HIV-1 subjects compared to healthy controls (data not shown).
Diagnostic potential of plasma miRNAs
ROC curve analysis was performed to evaluate the diagnostic potential of candidate miRNAs. The ROC curves of miR-29a, miR-223, miR-27a, miR-19b and miR-151-3p showed a high discrimination with AUC value of 0.949 (95% CI: 0.915-0.983), 0.905 (95% CI: 0.860–0.949), 0.897 (95% CI: 0.848–0.946), 0.989(95% CI: 0.974–1.003), 0.967(95% CI: 0.941–0.992), respectively (Figure 3). MiR-28-5p (95% CI: 0.703–0.859), miR-766 (95% CI: 0.694–0.842), and miR-30a-3p (95% CI: 0.680–0.834) showed a moderate discrimination with AUC value less than 0.8 (Figure 3). In order to increase the diagnostic efficiency of these markers, a combination of five miRNAs were used to show strong discrimination between the HIV-1 and control samples with high AUC value of 0.990 (Figure 4). Table 2 exhibits the specificity and sensitivity of each candidate miRNA with an optimal cutoff value. A cutoff value set at −6.00, the combined miRNAs showed a specificity of 97.3% and a sensitivity of 96.1% (Table 2).
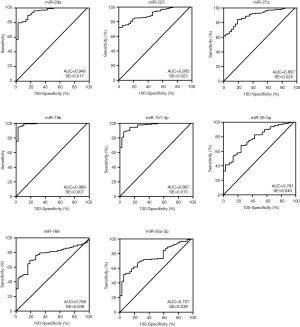
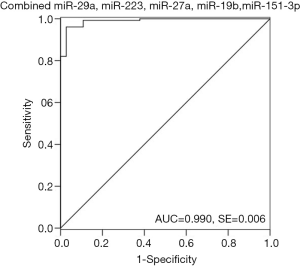
Table 2
| miRNA | Cutoff value | Sensitivity (%) | Specificity (%) |
|---|---|---|---|
| miR-29a | −7.13 | 79.7 | 97.3 |
| miR-223 | −2.42 | 71.9 | 100 |
| miR-27a | −6.37 | 84.4 | 81.1 |
| miR-19b | −5.43 | 94.5 | 97.3 |
| miR-151-3p | −5.41 | 88.3 | 94.6 |
| Combined miRNAs | −6.00 | 96.1 | 97.3 |
Different expression levels of eight miRNAs in LTC, MTC, and HTC groups
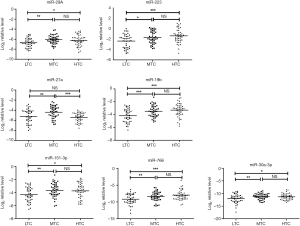
To monitor HIV/AIDS immune status, we further distinguished the expression levels of eight candidate miRNAs in LTC, MTC and HTC groups. The expression levels of miR-29a, miR-223, miR-27a, miR-19b, miR-151-3p, miR-766 and miR-30a-3p showed a significant difference among the LTC, MTC and HTC groups (P<0.05, ANOVA test). However, there was no significant difference in the expression levels of miR-28a-5p among these three groups (P>0.05, ANOVA test). Furthermore, multiple comparisons were carried out using the least significant difference (LSD) method. Figure 5 shows the different expression level of each miRNA in these three groups. Results from the comparison of the LTC and HTC groups indicated that six out of seven miRNAs (miR-29a, miR-151-3p, miR-223, miR-30a-3p, miR-19b, and miR-766) were significantly up-regulated (P<0.05, LSD test) in HTC group while miR-27a had no significant difference. The comparison between LTC and MTC groups showed that all the seven miRNAs were significantly up-regulated in MTC group (P<0.05, LSD test). While comparing MTC and HTC groups, only miR-27a showed a significant down-regulation in HTC group (P<0.001, LSD test). Altogether, our results showed that the expression levels of seven candidate miRNAs are significantly associated with CD4+ T cell count. Previous study showed that several host miRNAs play an important role in disease development (21-23)). For example, Patel et al. indicated that the miR-29a expression level was higher in PBMC and plasma from asymptomatic person (high CD4+ T cells) in whom virus replication is restricted, compared to symptomatic patients (low CD4+ T cells) in whom there is active viral replication (24)). Our results have shown that miR-29a, miR-223, miR-19b, miR-151-3p, miR-766 and miR-30a-3p have higher expression pattern in plasma from HIV-1 infected person with high CD4+ T cells compared to patients with low CD4+ T cells. Thus we hypothesize that the expression level of these six miRNAs may increase during HIV-1 latency and decrease during active viral replication although the mechanisms are still unknown. We noted that the baseline of CD4+ T cell count is very important. Compared with baseline CD4 <200 cell/µL, HIV-1 infected person with baseline CD4 >350 cell/µL or 200< CD4 ≤350 cell/µL showed similar expression pattern of above six miRNAs. A previous study has shown that patients with low level of CD4 + T cell count (CD4 <200 cell/µL) could impact immunological restoration (25). The CD4+ T cells are very important in forming immune response during HIV infection. So, our study showed that these six miRNAs could serve as useful biomarkers for monitoring the immune status of HIV/AIDS progression.
More and more researchers have attracted the attention of the role of miRNAs in pathogen-host interactions. Human miRNAs involved in many biological process, molecular function, and regulation pathways. During HIV-1 infections, miRNAs can either affect viral replication by targeting HIV-1 directly or modulate the expression of host genes and pathways essential for it by targeting host protein (26). For example, cellular miR-29a has been shown to inhibit HIV-1 replication by targeting the HIV-1 Nef transcripts (10,27). Also, cellular miR-28, miR-223, miR125b, miR-382 and miR-150 have been shown to be increased in resting primary CD4+ T cells compared to activated CD4+ T cells and can reduce HIV-1 replication by targeting HIV-1 mRNA transcripts (5). Our results also have shown a similar pattern of up-regulated miRNA expression such as miR-29a, miR-28 and miR-223 in plasma isolated from HIV-1 infected subjects. One study reported that miR-27a could decrease the phosphorylation of Akt and ERK, which could inhibit EV71 replication by targeting EGFR mRNA (28). The other study has demonstrated that miR-27a showed down-regulated in macrophages by targeting IL-10 through TLR2/4-driven inflammatory responses (29). Only 5% people known as HIV controllers can maintain high levels of T cells without antiretroviral therapy for more than 5 years (30). HIV/AIDS may cause serious complications including serious pneumonia, central nervous system complications, opportunistic infections and various viral-induced cancers (31,32). Previous studies have shown that over-expression of miR-19b and miR-151 was strongly associated with cancer invasion and metastasis (33-35). However, the mechanisms remain largely unknown. For the first time, our study has shown that higher level of miR-19b and miR-151-3p were expressed in HIV-1 patient plasma compared with healthy controls. Further studies are needed to reveal the function of them in HIV-1 infected subjects.
Conclusions
TLDA assays identified the differential expression of 231, 345, 315, and 257 miRNAs in plasma of healthy controls and HIV-1 LTC, MTC, and HTC groups, respectively. 5 miRNAs (miR-29a, miR-223, miR-27a, miR-151-3p and miR-19b) was combined as a useful biomarker for simple and efficient detection of HIV-1 infection. The biological mechanisms of these miRNAs need further investigation. Our results showed that the expression level of seven candidate miRNAs (miR-29a, miR-223, miR-766, miR-19b, miR-151-3p, miR-27a, and miR-30a-3p) were significantly associated with CD4+ T cell count and thus may serve as biomarkers for monitoring the HIV/AIDS immune progression. However, there are several limitations in our study. Firstly, not all of dysregulated miRNAs were confirmed in present study and other miRNAs might serve as more efficient biomarkers. Secondly, a larger numbers of subjects are required to confirm our study.
Table S1
| miRNA name | Raw Cq for control | Raw Cq for LTC | Fold change |
|---|---|---|---|
| hsa-miR-597-4380960 | 32.44 | 19.81 | 34,848.40 |
| hsa-miR-151-3p-002254 | 29.28 | 20.89 | 3,815.84 |
| hsa-miR-409-3p-002332 | 29.71 | 23.99 | 595.88 |
| hsa-miR-340#-002259 | 33.17 | 27.64 | 523.06 |
| hsa-miR-224-4395210 | 33.52 | 27.55 | 344.02 |
| hsa-miR-221-4373077 | 28.7 | 22.75 | 341.53 |
| hsa-miR-130b-4373144 | 34.81 | 28.88 | 334.19 |
| hsa-miR-766-001986 | 29.88 | 25 | 332.61 |
| hsa-miR-331-3p-4373046 | 30.03 | 24.15 | 325.20 |
| hsa-miR-425#-002302 | 33.36 | 28.58 | 310.21 |
| hsa-miR-339-5p-4395368 | 34.68 | 28.9 | 301.75 |
| hsa-miR-365-4373194 | 34.15 | 28.61 | 255.87 |
| hsa-miR-22#-002301 | 33.72 | 29.24 | 253.05 |
| hsa-miR-151-5P-002642 | 30.52 | 26.09 | 243.88 |
| hsa-miR-652-4395463 | 32.22 | 26.82 | 232.24 |
| hsa-miR-485-3p-4378095 | 32.32 | 27.01 | 218.12 |
| hsa-miR-125a-5p-4395309 | 34.83 | 29.53 | 215.81 |
| hsa-miR-30a-3p-000416 | 32.34 | 28.14 | 207.22 |
| hsa-miR-505#-002087 | 34.69 | 30.61 | 190.61 |
| hsa-miR-126#-000451 | 28.51 | 24.52 | 179.26 |
| hsa-miR-132-4373143 | 32.92 | 27.94 | 173.42 |
| hsa-miR-30e-3p-000422 | 29.21 | 25.31 | 168.84 |
| hsa-miR-15b-4373122 | 28.36 | 23.43 | 167.08 |
| hsa-miR-532-5p-4380928 | 32.1 | 27.2 | 164.14 |
| hsa-miR-199a-3p-4395415 | 28.5 | 23.6 | 163.67 |
| hsa-miR-625#-002432 | 30.39 | 26.58 | 158.85 |
| hsa-miR-425-4380926 | 34.69 | 29.9 | 151.73 |
| hsa-let-7d-4395394 | 28.97 | 24.2 | 150.12 |
| hsa-miR-324-5p-4373052 | 31.56 | 26.8 | 149.04 |
| hsa-miR-127-3p-4373147 | 30.89 | 26.22 | 139.78 |
| hsa-miR-584-001624 | 34.22 | 30.61 | 138.50 |
| hsa-miR-122-4395356 | 27.98 | 23.36 | 135.62 |
| hsa-miR-195-4373105 | 29.69 | 25.13 | 130.28 |
| hsa-miR-339-3p-4395295 | 33.87 | 29.38 | 123.93 |
| hsa-miR-27b-4373068 | 30.4 | 25.92 | 122.43 |
| hsa-miR-15a#-002419 | 34.61 | 31.19 | 121.47 |
| hsa-miR-28-5p-4373067 | 30.36 | 25.97 | 114.79 |
| hsa-miR-744-4395435 | 31.37 | 27.08 | 108.06 |
| hsa-miR-93#-002139 | 31.3 | 28.11 | 103.48 |
| hsa-miR-18a-4395533 | 29.5 | 25.29 | 102.32 |
| hsa-miR-24-4373072 | 24.87 | 20.66 | 102.03 |
| hsa-miR-382-4373019 | 33.77 | 29.57 | 101.70 |
| hsa-miR-491-5p-4381053 | 34.43 | 30.23 | 100.61 |
| hsa-miR-26a-4395166 | 27.34 | 23.15 | 100.33 |
| hsa-miR-192-4373108 | 32.4 | 28.24 | 98.52 |
| hsa-miR-25-4373071 | 27.4 | 23.27 | 96.02 |
| hsa-miR-495-4381078 | 31.25 | 27.14 | 95.05 |
| hsa-let-7g-4395393 | 30.74 | 26.63 | 94.89 |
| hsa-miR-27a-4373287 | 28.51 | 24.41 | 94.20 |
| hsa-miR-598-4395179 | 33.9 | 29.84 | 91.38 |
| hsa-miR-148b-4373129 | 31.36 | 27.32 | 90.37 |
| hsa-miR-223#-002098 | 31.42 | 28.52 | 84.32 |
| hsa-let-7a-4373169 | 30.62 | 26.71 | 83.11 |
| hsa-miR-671-3p-4395433 | 33.95 | 30.04 | 82.84 |
| hsa-miR-328-4373049 | 28.62 | 24.71 | 82.75 |
| hsa-miR-146a-4373132 | 25.85 | 21.96 | 81.67 |
| hsa-miR-574-3p-4395460 | 29.06 | 25.18 | 81.34 |
| hsa-miR-376c-4395233 | 32.17 | 28.31 | 80.02 |
| hsa-miR-222-4395387 | 27.45 | 23.62 | 78.40 |
| hsa-miR-191-4395410 | 25.37 | 21.54 | 78.28 |
| hsa-miR-376a-4373026 | 32.04 | 28.3 | 73.34 |
| hsa-miR-375-4373027 | 30.76 | 27.04 | 72.64 |
| hsa-miR-324-3p-4395272 | 32.99 | 29.3 | 70.89 |
| hsa-let-7b-4395446 | 30.12 | 26.45 | 70.13 |
| hsa-miR-301a-4373064 | 30.56 | 26.91 | 69.08 |
| hsa-miR-20a-4373286 | 26.01 | 22.39 | 67.56 |
| hsa-miR-125b-4373148 | 33.23 | 29.67 | 64.54 |
| hsa-miR-145-4395389 | 28.95 | 25.4 | 64.43 |
| hsa-miR-140-5p-4373374 | 29.86 | 26.35 | 62.83 |
| hsa-miR-885-5p-4395407 | 30.48 | 26.97 | 62.74 |
| hsa-miR-148a-4373130 | 29.46 | 25.97 | 61.77 |
| hsa-miR-142-3p-4373136 | 26.95 | 23.46 | 61.75 |
| hsa-miR-335-4373045 | 28.85 | 25.39 | 60.57 |
| hsa-miR-345-4395297 | 31.22 | 27.79 | 59.48 |
| hsa-miR-30d-000420 | 28.73 | 26.34 | 59.11 |
| hsa-let-7e-4395517 | 28 | 24.65 | 55.98 |
| hsa-miR-30a-5p-000417 | 25.2 | 22.93 | 54.81 |
| hsa-miR-93-4373302 | 27.1 | 23.8 | 54.15 |
| hsa-miR-17-4395419 | 23.97 | 20.68 | 53.77 |
| hsa-miR-16-4373121 | 25.2 | 21.96 | 52.03 |
| hsa-miR-152-4395170 | 29.09 | 25.86 | 51.62 |
| hsa-miR-340-4395369 | 30.89 | 27.71 | 49.99 |
| hsa-miR-26b-4395167 | 29.11 | 25.93 | 49.94 |
| hsa-miR-99b-4373007 | 32.29 | 29.13 | 49.02 |
| hsa-miR-223-4395406 | 19.61 | 16.46 | 48.72 |
| hsa-miR-21-4373090 | 25.68 | 22.54 | 48.58 |
| hsa-miR-30b-4373290 | 26 | 22.87 | 48.03 |
| hsa-miR-197-4373102 | 31.67 | 28.56 | 47.48 |
| hsa-miR-139-5p-4395400 | 28.9 | 25.8 | 47.32 |
| hsa-miR-106a-4395280 | 24.08 | 20.99 | 47.13 |
| hsa-miR-185-4395382 | 31.18 | 28.11 | 46.30 |
| hsa-miR-374b-4381045 | 28.06 | 25 | 46.07 |
| hsa-miR-374a-4373028 | 30.06 | 27 | 45.75 |
| hsa-miR-19b-4373098 | 24.42 | 21.44 | 43.31 |
| hsa-miR-320B-002844 | 30.31 | 28.45 | 41.12 |
| hsa-miR-146b-5p-4373178 | 28.19 | 25.34 | 39.83 |
| hsa-miR-28-3p-4395557 | 28.87 | 26.11 | 37.16 |
| hsa-miR-590-5p-4395176 | 31.14 | 28.44 | 35.94 |
| hsa-miR-484-4381032 | 22.87 | 20.21 | 34.64 |
| hsa-miR-136-4373173 | 34.7 | 32.09 | 33.63 |
| hsa-miR-338-5P-002658 | 34.29 | 32.75 | 33.05 |
| hsa-miR-126-4395339 | 23.51 | 20.98 | 31.84 |
| hsa-miR-128-4395327 | 29.63 | 27.09 | 31.84 |
| hsa-miR-10a-4373153 | 32.97 | 30.51 | 30.37 |
| hsa-miR-20b-4373263 | 27.94 | 25.49 | 30.03 |
| hsa-miR-218-4373081 | 31.36 | 28.93 | 29.70 |
| hsa-miR-720-002895 | 26.34 | 24.94 | 29.67 |
| hsa-miR-106b-4373155 | 26.19 | 23.77 | 29.47 |
| hsa-miR-186-4395396 | 28.87 | 26.49 | 28.49 |
| hsa-miR-361-5p-4373035 | 32.86 | 30.49 | 28.42 |
| hsa-miR-30c-4373060 | 24.52 | 22.19 | 27.78 |
| hsa-miR-92a-4395169 | 23.58 | 21.25 | 27.74 |
| hsa-miR-10b#-002315 | 34.54 | 33.26 | 27.44 |
| hsa-miR-454-4395434 | 31.3 | 28.99 | 27.42 |
| hsa-miR-101-4395364 | 31.44 | 29.14 | 27.19 |
| hsa-miR-194-4373106 | 33.45 | 31.17 | 26.74 |
| hsa-miR-29a-4395223 | 29.06 | 26.8 | 26.43 |
| hsa-miR-642-4380995 | 34.17 | 31.91 | 26.38 |
| hsa-miR-660-4380925 | 30.17 | 27.91 | 26.35 |
| hsa-miR-9#-002231 | 32.02 | 30.84 | 25.54 |
| hsa-miR-451-4373360 | 24.3 | 22.19 | 23.65 |
| hsa-miR-200c-4395411 | 34.73 | 32.67 | 23.03 |
| hsa-miR-378-002243 | 30.22 | 29.2 | 22.99 |
| hsa-miR-423-5p-4395451 | 31.26 | 29.21 | 22.74 |
| hsa-miR-193b-4395478 | 32.64 | 30.63 | 22.13 |
| hsa-miR-204-4373094 | 32.41 | 30.49 | 20.75 |
| hsa-miR-744#-002325 | 32.75 | 31.97 | 19.33 |
| hsa-miR-133a-4395357 | 30.1 | 28.34 | 18.71 |
| hsa-miR-579-4395509 | 32.89 | 31.18 | 18.02 |
| hsa-miR-1274B-002884 | 22.62 | 22.02 | 17.14 |
| hsa-miR-1290-002863 | 31.62 | 31.06 | 16.73 |
| hsa-miR-320-4395388 | 25.31 | 23.74 | 16.40 |
| hsa-miR-19a-4373099 | 28.74 | 27.25 | 15.48 |
| hsa-miR-362-3p-4395228 | 33.86 | 32.41 | 14.98 |
| hsa-miR-144#-002148 | 30 | 29.73 | 13.67 |
| hsa-miR-193a-5p-4395392 | 29.99 | 28.73 | 13.20 |
| hsa-miR-181a-4373117 | 33.57 | 32.49 | 11.64 |
| hsa-miR-337-5p-4395267 | 34.72 | 33.68 | 11.32 |
| hsa-miR-483-5p-4395449 | 30.29 | 29.29 | 10.94 |
| hsa-miR-342-3p-4395371 | 27.61 | 26.67 | 10.50 |
| hsa-miR-1255B-002801 | 32.51 | 33.56 | 5.48 |
| hsa-miR-367-4373034 | 31.01 | 31.05 | 5.37 |
| hsa-miR-150-4373127 | 24.56 | 24.72 | 4.94 |
| hsa-miR-486-5p-4378096 | 29.65 | 29.82 | 4.89 |
| hsa-miR-657-001512 | 33.43 | 34.73 | 4.60 |
| hsa-miR-381-4373020 | 20.47 | 20.92 | 4.04 |
| hsa-miR-100-4373160 | 30.51 | 31.53 | 2.71 |
| hsa-miR-661-001606 | 28.01 | 34.7 | 0.11 |
| hsa-miR-1197-002810 | 23.45 | 33.87 | 0.01 |
| hsa-miR-1274A-002883 | 12.73 | 27.43 | 0.00 |
The Fold change between two groups was calculated by 2−ΔΔCq method: ΔΔCq = ΔCqHIV infection − ΔCqcontrol; ΔCq = Cqtarget miRNA − CqU6. Italic labeled miRNAs were used for RT-qPCR confirmation test.
Table S2
| miRNA name | Raw Cq for control | Raw Cq for MTC | Fold change |
|---|---|---|---|
| hsa-miR-151-3p-002254 | 29.28 | 24.17 | 413.37 |
| hsa-miR-125a-5p-4395309 | 34.83 | 28.16 | 295.93 |
| hsa-miR-340#-002259 | 33.17 | 29.23 | 183.29 |
| hsa-miR-409-3p-002332 | 29.71 | 25.94 | 163.87 |
| hsa-miR-130b-4373144 | 34.81 | 29.31 | 131.40 |
| hsa-miR-151-5P-002642 | 30.52 | 27.16 | 122.80 |
| hsa-miR-10b#-002315 | 34.54 | 31.24 | 117.62 |
| hsa-miR-126#-000451 | 28.51 | 25.23 | 115.64 |
| hsa-miR-22#-002301 | 33.72 | 30.52 | 110.04 |
| hsa-miR-766-001986 | 29.88 | 26.69 | 109.48 |
| hsa-miR-30a-3p-000416 | 32.34 | 29.22 | 103.56 |
| hsa-miR-625#-002432 | 30.39 | 27.28 | 103.31 |
| hsa-miR-584-001624 | 34.22 | 31.18 | 98.65 |
| hsa-miR-365-4373194 | 34.15 | 29.1 | 96.27 |
| hsa-miR-331-3p-4373046 | 30.03 | 25.01 | 94.76 |
| hsa-miR-192-4373108 | 32.4 | 27.45 | 90.12 |
| hsa-miR-224-4395210 | 33.52 | 28.63 | 86.32 |
| hsa-miR-532-5p-4380928 | 32.1 | 27.24 | 84.96 |
| hsa-miR-15a#-002419 | 34.61 | 31.95 | 75.84 |
| hsa-miR-339-5p-4395368 | 34.68 | 29.99 | 75.17 |
| hsa-miR-132-4373143 | 32.92 | 28.47 | 63.35 |
| hsa-miR-425#-002302 | 33.36 | 31.02 | 60.52 |
| hsa-miR-652-4395463 | 32.22 | 27.85 | 60.05 |
| hsa-miR-30e-3p-000422 | 29.21 | 26.95 | 57.30 |
| hsa-miR-30a-5p-000417 | 25.2 | 23.08 | 52.07 |
| hsa-miR-221-4373077 | 28.7 | 24.55 | 51.60 |
| hsa-miR-199a-3p-4395415 | 28.5 | 24.36 | 50.98 |
| hsa-miR-25-4373071 | 27.4 | 23.28 | 50.37 |
| hsa-miR-18a-4395533 | 29.5 | 25.44 | 48.72 |
| hsa-miR-495-4381078 | 31.25 | 27.2 | 48.03 |
| hsa-miR-30d-000420 | 28.73 | 26.74 | 47.40 |
| hsa-miR-485-3p-4378095 | 32.32 | 28.33 | 46.21 |
| hsa-miR-1243-002854 | 19.76 | 17.84 | 45.34 |
| hsa-miR-15b-4373122 | 28.36 | 24.46 | 43.15 |
| hsa-miR-324-3p-4395272 | 32.99 | 29.12 | 42.48 |
| hsa-miR-375-4373027 | 30.76 | 26.91 | 42.15 |
| hsa-miR-598-4395179 | 33.9 | 30.06 | 41.42 |
| hsa-miR-324-5p-4373052 | 31.56 | 27.78 | 39.92 |
| hsa-miR-195-4373105 | 29.69 | 25.92 | 39.80 |
| hsa-miR-320B-002844 | 30.31 | 28.6 | 39.03 |
| hsa-miR-24-4373072 | 24.87 | 21.13 | 38.74 |
| hsa-miR-93#-002139 | 31.3 | 29.61 | 38.69 |
| hsa-miR-19b-4373098 | 24.42 | 20.69 | 38.41 |
| hsa-miR-223#-002098 | 31.42 | 29.79 | 37.04 |
| hsa-miR-339-3p-4395295 | 33.87 | 30.25 | 35.94 |
| hsa-miR-27a-4373287 | 28.51 | 24.91 | 35.41 |
| hsa-miR-491-5p-4381053 | 34.43 | 30.85 | 34.80 |
| hsa-miR-671-3p-4395433 | 33.95 | 30.39 | 34.35 |
| hsa-miR-28-5p-4373067 | 30.36 | 26.8 | 34.29 |
| hsa-miR-16-4373121 | 25.2 | 21.66 | 33.78 |
| hsa-miR-505#-002087 | 34.69 | 33.2 | 33.67 |
| hsa-miR-642-4380995 | 34.17 | 30.65 | 33.51 |
| hsa-miR-26a-4395166 | 27.34 | 23.83 | 33.24 |
| hsa-miR-20a-4373286 | 26.01 | 22.58 | 31.20 |
| hsa-miR-338-5P-002658 | 34.29 | 32.93 | 30.80 |
| hsa-miR-26b-4395167 | 29.11 | 25.75 | 29.92 |
| hsa-miR-125b-4373148 | 33.23 | 29.89 | 29.31 |
| hsa-let-7d-4395394 | 28.97 | 25.64 | 29.27 |
| hsa-miR-144#-002148 | 30 | 28.72 | 29.20 |
| hsa-miR-140-5p-4373374 | 29.86 | 26.54 | 29.02 |
| hsa-miR-145-4395389 | 28.95 | 25.63 | 29.00 |
| hsa-let-7g-4395393 | 30.74 | 27.43 | 28.90 |
| hsa-miR-660-4380925 | 30.17 | 26.87 | 28.57 |
| hsa-miR-142-3p-4373136 | 26.95 | 23.68 | 27.96 |
| hsa-miR-136-4373173 | 34.7 | 31.45 | 27.73 |
| hsa-miR-127-3p-4373147 | 30.89 | 27.72 | 26.17 |
| hsa-miR-328-4373049 | 28.62 | 25.47 | 25.79 |
| hsa-miR-374a-4373028 | 30.06 | 26.93 | 25.34 |
| hsa-miR-93-4373302 | 27.1 | 23.97 | 25.32 |
| hsa-miR-27b-4373068 | 30.4 | 27.3 | 24.83 |
| hsa-miR-185-4395382 | 31.18 | 28.13 | 24.07 |
| hsa-miR-345-4395297 | 31.22 | 28.19 | 23.75 |
| hsa-miR-222-4395387 | 27.45 | 24.44 | 23.38 |
| hsa-miR-146a-4373132 | 25.85 | 22.84 | 23.37 |
| hsa-miR-122-4395356 | 27.98 | 24.98 | 23.32 |
| hsa-miR-148a-4373130 | 29.46 | 26.47 | 23.13 |
| hsa-miR-744-4395435 | 31.37 | 28.39 | 22.94 |
| hsa-miR-376c-4395233 | 32.17 | 29.21 | 22.68 |
| hsa-miR-21-4373090 | 25.68 | 22.72 | 22.63 |
| hsa-miR-301a-4373064 | 30.56 | 27.64 | 21.93 |
| hsa-miR-148b-4373129 | 31.36 | 28.46 | 21.69 |
| hsa-miR-181a-4373117 | 33.57 | 30.68 | 21.59 |
| hsa-miR-376a-4373026 | 32.04 | 29.2 | 20.83 |
| hsa-miR-451-4373360 | 24.3 | 21.47 | 20.65 |
| hsa-miR-191-4395410 | 25.37 | 22.57 | 20.18 |
| hsa-miR-340-4395369 | 30.89 | 28.14 | 19.58 |
| hsa-miR-30b-4373290 | 26 | 23.3 | 18.91 |
| hsa-miR-378-002243 | 30.22 | 29.57 | 18.77 |
| hsa-miR-590-5p-4395176 | 31.14 | 28.46 | 18.64 |
| hsa-miR-99b-4373007 | 32.29 | 29.62 | 18.52 |
| hsa-miR-19a-4373099 | 28.74 | 26.07 | 18.51 |
| hsa-miR-744#-002325 | 32.75 | 32.15 | 18.08 |
| hsa-miR-223-4395406 | 19.61 | 17.06 | 17.01 |
| hsa-miR-92a-4395169 | 23.58 | 21.03 | 16.98 |
| hsa-miR-106a-4395280 | 24.08 | 21.54 | 16.94 |
| hsa-miR-17-4395419 | 23.97 | 21.44 | 16.73 |
| hsa-miR-20b-4373263 | 27.94 | 25.44 | 16.42 |
| hsa-miR-425-4380926 | 34.69 | 32.19 | 16.37 |
| hsa-miR-361-5p-4373035 | 32.86 | 30.39 | 16.18 |
| hsa-miR-146b-5p-4373178 | 28.19 | 25.73 | 15.98 |
| hsa-miR-335-4373045 | 28.85 | 26.42 | 15.70 |
| hsa-let-7e-4395517 | 28 | 25.57 | 15.62 |
| hsa-miR-197-4373102 | 31.67 | 29.27 | 15.40 |
| hsa-miR-885-5p-4395407 | 30.48 | 28.08 | 15.37 |
| hsa-miR-454-4395434 | 31.3 | 28.92 | 15.18 |
| hsa-miR-374b-4381045 | 28.06 | 25.69 | 15.07 |
| hsa-miR-139-5p-4395400 | 28.9 | 26.57 | 14.65 |
| hsa-miR-101-4395364 | 31.44 | 29.12 | 14.55 |
| hsa-miR-29a-4395223 | 29.06 | 26.76 | 14.33 |
| hsa-miR-582-3p-4395510 | 34.91 | 32.62 | 14.24 |
| hsa-let-7b-4395446 | 30.12 | 27.84 | 14.16 |
| hsa-miR-10a-4373153 | 32.97 | 30.76 | 13.46 |
| hsa-miR-106b-4373155 | 26.19 | 24.01 | 13.13 |
| hsa-miR-579-4395509 | 32.89 | 30.73 | 13.04 |
| hsa-miR-126-4395339 | 23.51 | 21.38 | 12.77 |
| hsa-miR-484-4381032 | 22.87 | 20.76 | 12.49 |
| hsa-miR-128-4395327 | 29.63 | 27.53 | 12.40 |
| hsa-miR-574-3p-4395460 | 29.06 | 26.98 | 12.37 |
| hsa-miR-194-4373106 | 33.45 | 31.37 | 12.26 |
| hsa-let-7a-4373169 | 30.62 | 28.59 | 11.89 |
| hsa-miR-186-4395396 | 28.87 | 26.92 | 11.19 |
| hsa-miR-152-4395170 | 29.09 | 27.16 | 11.09 |
| hsa-miR-382-4373019 | 33.77 | 31.88 | 10.84 |
| hsa-miR-193a-5p-4395392 | 29.99 | 28.1 | 10.76 |
| hsa-miR-30c-4373060 | 24.52 | 22.65 | 10.63 |
| hsa-miR-1274B-002884 | 22.62 | 22.85 | 10.19 |
| hsa-miR-193b-4395478 | 32.64 | 30.91 | 9.62 |
| hsa-miR-9#-002231 | 32.02 | 32.45 | 8.86 |
| hsa-miR-28-3p-4395557 | 28.87 | 27.32 | 8.49 |
| hsa-miR-1290-002863 | 31.62 | 32.28 | 7.59 |
| hsa-miR-720-002895 | 26.34 | 27.02 | 7.45 |
| hsa-miR-423-5p-4395451 | 31.26 | 29.93 | 7.28 |
| hsa-miR-483-5p-4395449 | 30.29 | 28.98 | 7.17 |
| hsa-miR-342-3p-4395371 | 27.61 | 26.34 | 6.98 |
| hsa-miR-367-4373034 | 31.01 | 29.77 | 6.87 |
| hsa-miR-16-1#-002420 | 33.63 | 34.52 | 6.49 |
| hsa-miR-320-4395388 | 25.31 | 24.16 | 6.44 |
| hsa-miR-150-4373127 | 24.56 | 23.46 | 6.23 |
| hsa-miR-486-5p-4378096 | 29.65 | 28.64 | 5.84 |
| hsa-miR-601-001558 | 32.97 | 34.04 | 5.69 |
| hsa-miR-362-3p-4395228 | 33.86 | 32.9 | 5.66 |
| hsa-miR-200c-4395411 | 34.73 | 33.89 | 5.21 |
| hsa-miR-133a-4395357 | 30.1 | 29.39 | 4.76 |
| hsa-miR-204-4373094 | 32.41 | 32.1 | 3.61 |
| hsa-miR-1255B-002801 | 32.51 | 34.32 | 3.41 |
| hsa-miR-661-001606 | 28.01 | 30.09 | 2.83 |
| hsa-miR-381-4373020 | 20.47 | 20.77 | 2.37 |
| hsa-miR-218-4373081 | 31.36 | 31.88 | 2.03 |
| has-miR-155-4395459 | 24.15 | 26.9 | 0.43 |
| hsa-miR-1274A-002883 | 12.73 | 28.92 | 0.00 |
The Fold change between two groups was calculated by 2−ΔΔCq method: ΔΔCq =ΔCqHIV infectiont − ΔCqcontrol; ΔCq = Cqtarget miRNA − CqU6. Italic labeled miRNAs were used for RT-qPCR confirmation test.
Table S3
| miRNA name | Raw Cq for control | Raw Cq for HTC | Fold change |
|---|---|---|---|
| hsa-miR-151-3p-002254 | 29.28 | 22.02 | 2,539.61 |
| hsa-miR-766-001986 | 29.88 | 26.92 | 129.22 |
| hsa-miR-1243-002854 | 19.76 | 16.85 | 124.63 |
| hsa-miR-340#-002259 | 33.17 | 30.78 | 86.48 |
| hsa-miR-505#-002087 | 34.69 | 32.55 | 73.16 |
| hsa-miR-409-3p-002332 | 29.71 | 27.71 | 66.10 |
| hsa-miR-625#-002432 | 30.39 | 28.72 | 52.72 |
| hsa-miR-338-5P-002658 | 34.29 | 32.92 | 42.88 |
| hsa-miR-425#-002302 | 33.36 | 32.31 | 34.29 |
| hsa-miR-22#-002301 | 33.72 | 32.69 | 33.80 |
| hsa-miR-126#-000451 | 28.51 | 27.51 | 33.11 |
| hsa-miR-224-4395210 | 33.52 | 30.2 | 27.81 |
| hsa-miR-151-5P-002642 | 30.52 | 29.82 | 26.94 |
| hsa-miR-365-4373194 | 34.15 | 31.1 | 23.11 |
| hsa-miR-125a-5p-4395309 | 34.83 | 31.81 | 22.58 |
| hsa-miR-30e-3p-000422 | 29.21 | 28.87 | 20.93 |
| hsa-miR-320B-002844 | 30.31 | 30.18 | 18.15 |
| hsa-let-7b-4395446 | 30.12 | 27.5 | 17.15 |
| hsa-miR-485-3p-4378095 | 32.32 | 29.7 | 17.09 |
| hsa-miR-367-4373034 | 31.01 | 28.65 | 14.38 |
| hsa-miR-328-4373049 | 28.62 | 26.27 | 14.22 |
| hsa-miR-30a-3p-000416 | 32.34 | 32.58 | 14.01 |
| hsa-miR-518b-4373246 | 31.67 | 29.4 | 13.51 |
| hsa-miR-25-4373071 | 27.4 | 25.17 | 13.08 |
| hsa-miR-324-5p-4373052 | 31.56 | 29.35 | 12.94 |
| hsa-miR-221-4373077 | 28.7 | 26.51 | 12.75 |
| hsa-miR-192-4373108 | 32.4 | 30.25 | 12.39 |
| hsa-miR-324-3p-4395272 | 32.99 | 30.89 | 11.95 |
| hsa-miR-671-3p-4395433 | 33.95 | 31.93 | 11.34 |
| hsa-miR-30a-5p-000417 | 25.2 | 25.75 | 11.32 |
| hsa-miR-652-4395463 | 32.22 | 30.2 | 11.29 |
| hsa-miR-199a-3p-4395415 | 28.5 | 26.59 | 10.43 |
| hsa-miR-331-3p-4373046 | 30.03 | 28.2 | 9.95 |
| hsa-miR-28-5p-4373067 | 30.36 | 28.55 | 9.74 |
| hsa-let-7d-4395394 | 28.97 | 27.18 | 9.69 |
| hsa-miR-375-4373027 | 30.76 | 28.99 | 9.53 |
| hsa-miR-145-4395389 | 28.95 | 27.19 | 9.42 |
| hsa-miR-223#-002098 | 31.42 | 32.3 | 9.03 |
| hsa-miR-744-4395435 | 31.37 | 29.7 | 8.87 |
| hsa-miR-598-4395179 | 33.9 | 32.26 | 8.69 |
| hsa-miR-93#-002139 | 31.3 | 32.27 | 8.47 |
| hsa-miR-144#-002148 | 30 | 30.98 | 8.43 |
| hsa-miR-127-3p-4373147 | 30.89 | 29.34 | 8.19 |
| hsa-miR-136-4373173 | 34.7 | 33.19 | 7.97 |
| hsa-miR-122-4395356 | 27.98 | 26.47 | 7.96 |
| hsa-miR-125b-4373148 | 33.23 | 31.72 | 7.92 |
| hsa-let-7e-4395517 | 28 | 26.5 | 7.87 |
| hsa-let-7g-4395393 | 30.74 | 29.25 | 7.83 |
| hsa-miR-18a-4395533 | 29.5 | 28.04 | 7.69 |
| hsa-miR-197-4373102 | 31.67 | 30.24 | 7.53 |
| hsa-miR-1274B-002884 | 22.62 | 23.77 | 7.46 |
| hsa-miR-484-4381032 | 22.87 | 21.45 | 7.46 |
| hsa-miR-195-4373105 | 29.69 | 28.33 | 7.15 |
| hsa-miR-376a-4373026 | 32.04 | 30.69 | 7.11 |
| hsa-miR-495-4381078 | 31.25 | 29.95 | 6.85 |
| hsa-miR-491-5p-4381053 | 34.43 | 33.22 | 6.44 |
| hsa-miR-92a-4395169 | 23.58 | 22.38 | 6.42 |
| hsa-miR-133a-4395357 | 30.1 | 28.91 | 6.36 |
| hsa-miR-142-3p-4373136 | 26.95 | 25.8 | 6.18 |
| hsa-miR-26a-4395166 | 27.34 | 26.22 | 6.08 |
| hsa-miR-15b-4373122 | 28.36 | 27.24 | 6.02 |
| hsa-miR-657-001512 | 33.43 | 34.9 | 5.96 |
| hsa-miR-885-5p-4395407 | 30.48 | 29.39 | 5.94 |
| hsa-miR-454-4395434 | 31.3 | 30.22 | 5.91 |
| hsa-miR-130b-4373144 | 34.81 | 33.77 | 5.73 |
| hsa-miR-30d-000420 | 28.73 | 30.3 | 5.56 |
| hsa-miR-10a-4373153 | 32.97 | 31.98 | 5.55 |
| hsa-miR-93-4373302 | 27.1 | 26.14 | 5.41 |
| hsa-miR-532-5p-4380928 | 32.1 | 31.21 | 5.19 |
| hsa-miR-1290-002863 | 31.62 | 33.3 | 5.17 |
| hsa-miR-486-5p-4378096 | 29.65 | 28.78 | 5.10 |
| hsa-miR-574-3p-4395460 | 29.06 | 28.2 | 5.09 |
| hsa-miR-24-4373072 | 24.87 | 24 | 5.09 |
| hsa-miR-381-4373020 | 20.47 | 19.63 | 4.98 |
| hsa-let-7a-4373169 | 30.62 | 29.79 | 4.97 |
| hsa-miR-222-4395387 | 27.45 | 26.63 | 4.92 |
| hsa-miR-361-5p-4373035 | 32.86 | 32.08 | 4.81 |
| hsa-miR-720-002895 | 26.34 | 28.14 | 4.73 |
| hsa-miR-339-5p-4395368 | 34.68 | 33.93 | 4.68 |
| hsa-miR-193b-4395478 | 32.64 | 31.9 | 4.66 |
| hsa-miR-27a-4373287 | 28.51 | 27.85 | 4.42 |
| hsa-miR-660-4380925 | 30.17 | 29.51 | 4.40 |
| hsa-miR-26b-4395167 | 29.11 | 28.49 | 4.31 |
| hsa-miR-16-4373121 | 25.2 | 24.6 | 4.23 |
| hsa-miR-30b-4373290 | 26 | 25.43 | 4.14 |
| hsa-miR-642-4380995 | 34.17 | 33.65 | 4.00 |
| hsa-miR-378-002243 | 30.22 | 32.28 | 3.98 |
| hsa-miR-423-5p-4395451 | 31.26 | 30.85 | 3.70 |
| hsa-miR-20a-4373286 | 26.01 | 25.61 | 3.67 |
| hsa-miR-193a-5p-4395392 | 29.99 | 29.61 | 3.62 |
| hsa-miR-302c-4378072 | 19.6 | 19.26 | 3.52 |
| hsa-miR-27b-4373068 | 30.4 | 30.08 | 3.47 |
| hsa-miR-345-4395297 | 31.22 | 30.93 | 3.41 |
| hsa-miR-382-4373019 | 33.77 | 33.49 | 3.39 |
| hsa-miR-146a-4373132 | 25.85 | 25.6 | 3.30 |
| hsa-miR-99b-4373007 | 32.29 | 32.05 | 3.29 |
| hsa-miR-28-3p-4395557 | 28.87 | 28.64 | 3.25 |
| hsa-miR-181a-4373117 | 33.57 | 33.44 | 3.06 |
| hsa-miR-374a-4373028 | 30.06 | 29.95 | 3.02 |
| hsa-miR-19b-4373098 | 24.42 | 24.33 | 2.97 |
| hsa-miR-191-4395410 | 25.37 | 25.32 | 2.88 |
| hsa-miR-148a-4373130 | 29.46 | 29.44 | 2.84 |
| hsa-miR-223-4395406 | 19.61 | 19.6 | 2.81 |
| hsa-miR-128-4395327 | 29.63 | 29.63 | 2.78 |
| hsa-miR-152-4395170 | 29.09 | 29.12 | 2.73 |
| hsa-miR-422a-4395408 | 34.78 | 34.82 | 2.71 |
| hsa-miR-451-4373360 | 24.3 | 24.36 | 2.66 |
| hsa-miR-150-4373127 | 24.56 | 24.63 | 2.66 |
| hsa-miR-21-4373090 | 25.68 | 25.78 | 2.60 |
| hsa-miR-139-5p-4395400 | 28.9 | 29.17 | 2.32 |
| hsa-miR-30c-4373060 | 24.52 | 24.81 | 2.29 |
| hsa-miR-301a-4373064 | 30.56 | 30.87 | 2.25 |
| hsa-miR-140-5p-4373374 | 29.86 | 30.2 | 2.21 |
| hsa-miR-106a-4395280 | 24.08 | 24.49 | 2.10 |
| hsa-miR-374b-4381045 | 28.06 | 28.5 | 2.05 |
| has-miR-155-4395459 | 24.15 | 27.64 | 0.25 |
| hsa-miR-1274A-002883 | 12.73 | 33.07 | 0.00 |
The Fold change between two groups was calculated by 2−ΔΔCq method: ΔΔCq =ΔCqHIV infection − ΔCqcontrol; ΔCq = Cqtarget miRNA − CqU6. Italic labeled miRNAs were used for RT-qPCR confirmation test.
Acknowledgments
The authors thank Doulathunnisa Ali help to correct the manuscript and thank the patients who participated in our study.
Funding: This work was supported by the Natural Science Foundation of Jiangsu Province (bk20131451, bk20161583andbk20141030) and Jiangsu Province Science & Technology Demonstration Project for Emerging Infectious Diseases Control and Prevention (BE2015714).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jphe.2017.05.11). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study has been approved by the Ethics Committee of Jiangsu Provincial Center for Diseases Prevention and Control (approval number: JSCDCLL2013014). The written informed consent was obtained from all participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- 2015 China AIDS Response Progress Report. Available online: http://www.aidsdatahub.org/sites/default/files/publication/China_narrative_report_2015.pdf
- Constantine NT, Kabat W, Zhao RY. Update on the laboratory diagnosis and monitoring of HIV infection. Cell Res 2005;15:870-6. [Crossref] [PubMed]
- Grassmann R, Jeang KT. The roles of microRNAs in mammalian virus infection. Biochim Biophys Acta 2008;1779:706-11.
- Catalucci D, Latronico MV, Condorelli G. MicroRNAs control gene expression: importance for cardiac development and pathophysiology. Ann N Y Acad Sci 2008;1123:20-9. [Crossref] [PubMed]
- Huang J, Wang F, Argyris E, et al. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med 2007;13:1241-7. [Crossref] [PubMed]
- Wang X, Ye L, Hou W, et al. Cellular microRNA expression correlates with susceptibility of monocytes/macrophages to HIV-1 infection. Blood 2009;113:671-4. [Crossref] [PubMed]
- Wu JQ, Dwyer DE, Dyer WB, et al. Genome-wide analysis of primary CD4+ and CD8+ T cell transcriptomes shows evidence for a network of enriched pathways associated with HIV disease. Retrovirology 2011;8:18. [Crossref] [PubMed]
- Witwer KW, Watson AK, Blankson JN, et al. Relationships of PBMC microRNA expression, plasma viral load, and CD4+ T-cell count in HIV-1-infected elite suppressors and viremic patients. Retrovirology 2012;9:5. [Crossref] [PubMed]
- Bignami F, Pilotti E, Bertoncelli L, et al. Stable changes in CD4+ T lymphocyte miRNA expression after exposure to HIV-1. Blood 2012;119:6259-67. [Crossref] [PubMed]
- Ahluwalia JK, Khan SZ, Soni K, et al. Human cellular microRNA hsa-miR-29a interferes with viral nef protein expression and HIV-1 replication. Retrovirology 2008;5:117. [Crossref] [PubMed]
- Godfrey AC, Xu Z, Weinberg CR, et al. Serum microRNA expression as an early marker for breast cancer risk in prospectively collected samples from the Sister Study cohort. Breast Cancer Res 2013;15:R42. [Crossref] [PubMed]
- Hennessey PT, Sanford T, Choudhary A, et al. Serum microRNA biomarkers for detection of non-small cell lung cancer. Plos One 2012;7:e32307 [Crossref] [PubMed]
- Cui L, Qi Y, Li H, et al. Serum microRNA expression profile distinguishes enterovirus 71 and coxsackievirus 16 infections in patients with hand-foot-and-mouth disease. PLoS One 2011;6:e27071 [Crossref] [PubMed]
- Sun Q, Zhang J, Cao W, et al. Dysregulated miR-363 affects head and neck cancer invasion and metastasis by targeting podoplanin. Int J Biochem Cell Biol 2013;45:513-20. [Crossref] [PubMed]
- Qi Y, Cui L, Ge Y, et al. Altered serum microRNAs as biomarkers for the early diagnosis of pulmonary tuberculosis infection. BMC Infect Dis 2012;12:384. [Crossref] [PubMed]
- Zhu Z, Qi Y, Ge A, et al. Comprehensive characterization of serum microRNA profile in response to the emerging avian influenza A (H7N9) virus infection in humans. Viruses 2014;6:1525-39. [Crossref] [PubMed]
- Chen X, Ba Y, Ma LJ, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Research 2008;18:997-1006. [Crossref] [PubMed]
- Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513-8. [Crossref] [PubMed]
- Qi Y, Zhu Z, Shi Z, et al. Dysregulated microRNA expression in serum of non-vaccinated children with varicella. Viruses 2014;6:1823-36. [Crossref] [PubMed]
- Redell JB, Moore AN, Ward NH 3rd, et al. Human traumatic brain injury alters plasma microRNA levels. J Neurotrauma 2010;27:2147-56. [Crossref] [PubMed]
- Duskova K, Nagilla P, Le HS, et al. MicroRNA regulation and its effects on cellular transcriptome in human immunodeficiency virus-1 (HIV-1) infected individuals with distinct viral load and CD4 cell counts. BMC Infect Dis 2013;13:250. [Crossref] [PubMed]
- Geiss GK, Bumgarner RE, An MC, et al. Large-scale monitoring of host cell gene expression during HIV-1 infection using cDNA microarrays. Virology 2000;266:8-16. [Crossref] [PubMed]
- Ryo A, Suzuki Y, Arai M, et al. Identification and characterization of differentially expressed mRNAs in HIV type 1-infected human T cells. AIDS Res Hum Retroviruses 2000;16:995-1005. [Crossref] [PubMed]
- Patel P, Ansari MY, Bapat S, et al. The microRNA miR-29a is associated with human immunodeficiency virus latency. Retrovirology 2014;11:108. [Crossref] [PubMed]
- Zhang F, Sun M, Sun J, et al. The risk factors for suboptimal CD4 recovery in HIV infected population: an observational and retrospective study in Shanghai, China. Biosci Trends 2015;9:335-41. [Crossref] [PubMed]
- Swaminathan S, Murray DD, Kelleher AD. miRNAs and HIV: unforeseen determinants of host-pathogen interaction. Immunol Rev 2013;254:265-80. [Crossref] [PubMed]
- Nathans R, Chu CY, Serquina AK, et al. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell 2009;34:696-709. [Crossref] [PubMed]
- Zhang L, Chen X, Shi Y, et al. miR-27a suppresses EV71 replication by directly targeting EGFR. Virus Genes 2014;49:373-82. [Crossref] [PubMed]
- Xie N, Cui H, Banerjee S, et al. miR-27a regulates inflammatory response of macrophages by targeting IL-10. J Immunol 2014;193:327-34. [Crossref] [PubMed]
- Blankson JN. Control of HIV-1 replication in elite suppressors. Discov Med 2010;9:261-6. [PubMed]
- Vogel M, Schwarze-Zander C, Wasmuth JC, et al. The treatment of patients with HIV. Dtsch Arztebl Int 2010;107:507-15; quiz 16. [PubMed]
- Chu C, Selwyn PA. Complications of HIV infection: a systems-based approach. Am Fam Physician 2011;83:395-406. [PubMed]
- Li J, Yang S, Yan W, et al. MicroRNA-19 triggers epithelial-mesenchymal transition of lung cancer cells accompanied by growth inhibition. Lab Invest 2015;95:1056-70. [Crossref] [PubMed]
- Luedde T. MicroRNA-151 and its hosting gene FAK (focal adhesion kinase) regulate tumor cell migration and spreading of hepatocellular carcinoma. Hepatology 2010;52:1164-6. [Crossref] [PubMed]
- Xiong Y, Kotian S, Zeiger MA, et al. miR-126-3p Inhibits Thyroid Cancer Cell Growth and Metastasis, and Is Associated with Aggressive Thyroid Cancer. Plos One 2015;10:e0130496 [Crossref] [PubMed]
Cite this article as: Qi Y, Hu H, Guo H, Xu P, Shi Z, Huan X, Zhu Z, Zhou M, Cui L. MicroRNA profiling in plasma of HIV-1 infected patients: potential markers of infection and immune status. J Public Health Emerg 2017;1:65.


