
Molecular evolution of the major capsid gene in human Norovirus GII.17
Introduction
Norovirus is a leading cause of acute gastroenteritis (AGE) globally, estimated to cause nearly 20% of all AGE cases worldwide (1-4). In the developing countries, AGE leading to dehydration and malnutrition primarily affects young children (3,5,6). Human Norovirus (HuNoV) belongs to the genus Norovirus in the family Caliciviridae and, is classified into seven genogroups (GI–GVII) based on phylogenetic analysis of the major capsid gene (VP1). Of them, GI, GII, GIV may infect humans and GII genotype (e.g., GII.2, GII.3, GII.4 and GII.6) has ever caused AGE prevalence (7). Especially, GII.4 had caused four epidemics of AGE in history, and has been the predominant genotype among all HuNoV genotypes causing human infection (8,9). However, in recent years, GII.17 frequently emerges, and become one of the major genotypes causing AGE in some countries (10).
HuNoV VP1 protein is closely associated with the infectivity and antigenicity of these strains (11,12). Many previous reports showed that the HuNov VP1 gene rapidly evolved, resulting in a large divergence of antigenicity (13). Earlier findings also indicated that the rapid evolution of the VP1 gene of Norovirus is strongly associated with AGE epidemics caused by HuNoV. To better understand the characteristics of Norovirus GII.17 isolated globally, we analyzed positive and negative selective sites, similarity, and evolution of VP1 gene in this study.
Methods
Strains selection
The full-length sequences of GII.17 norovirus VP1 gene (1,623 bp) were downloaded from GenBank. Which included all sequences submitted as of September 14th, 2018. A total of 449 strains were obtained. These sequences were aligned using MEGA7.0 software with muscle program. All sequences were coded according to GenBank accession number/location of virus isolation/the year of virus isolation. Due to limitation in computing capacity, positive and negative selection analyses could not include sequences with greater than 99.0% identity, the VP1 sequences with >99.0% were excluded from this analysis. The identity of VP1 sequences were calculated based on gene distances of VP1 sequences which were analyzed using MEGA7.0 software. Only 38 sequences were selected to do dN/dS analysis and phylogenetic tree construction. The GenBank Assess No. are as follows: FJ537136, MF918359, KR154230, KU561251, KT326181, KJ156329, LC349991, LC148854, LC101820, KT285173, KX424647, KX171413, KX171415, KX171412, KX420894, KX171417, KX171418, KX171416, KT315673, KT315698, KT315706, KT315718, KU953397, KU561227, KU561242, KU561245, KU557808, KU557813, KX244854, KY069114, KX168439, KX168444, KY406957, KY406971, KY406974, KP902563, KP902565, KU557839, KU587625. The sequences of other GII genotype sequences were used as the reference strain. GenBank Assess No. are as follows: KM268102, KF306214, JQ622197, FJ537134, KY407196, KY406943, KY424341, KY424342, KY424346, KY424345, KY406940, KY457583.
Model selection analyses
The best-fit model for nucleotide substitution was used to compute likelihoods with jModelTest v3.7. The phylogenetic tree is constructed using a Bayesian Markov Chain Monte Carlo approach under the GTR model of nucleotide substitution with a proportion of invariable sites and substitution rate heterogeneity implemented in BEAST v1.8.4. The sequences were partitioned into 3-codon positions. The convergence of parameters was analyzed using Tracer v1.7.1. The effective sample size of each parameter calculated was above 200. The maximum clade credibility tree was generated with program TreeAnnotator using TreeAnnotator v1.8.4.
The distance of VP1 gene of norovirus GII.17 analysis
According to MCMC phylogenetic tree, VP1 gene sequences of GII.17 was labeled group 1 and 2, respectively. The gene distance of VP1 within various clades and between clades were calculated using distance program with MEGA7.0 software.
Selective pressure analysis
The ration of Nonsynonymous(dN) to synonymous(dS) substitutions at every codon were calculated online using Datamonkey Adaptive Evolution Server (http://www.datamonkey.org/). FEL, and MEME methods were used. The dN/dS ration was estimated under the MG94 model in the Datamonkey. The cut off P value was at ≤0.05.
Similarity analysis
All 38 sequence of VP1 gene were used to do similarity analysis using Simplot software. MF918359 was used as query sequences, KR154230 and KU561251 as reference sequences. The rest of 35 sequences were all included. The similarity was examined using a window size of 200 nucleotides in length (nt) and a step size of 20 nt in the full-length VP1 genes.
Results
Time-scale evolution of the globally collected GII.17 strains
MCMC phylogenetic tree was constructed based on the full-length capsid gene (shown in Figure 1). All Norovirus GII.17 was classified into two clades. The MCMC trees showed that the most recent common ancestor of GII.17 was around 1984.6 (1926.9–1995.0). The mean evolutionary rate of the present human GII.17 strains was estimated to be 2.31×10−3 substitutions/site/year [95% highest posterior densities (HPDs) 9.40×10−4–3.84×10−3 substitutions/site/year].

Genetic diversity of GII.17
The mean gene distance is 0.012 (95% CI: 0.011–0.013) within cluster 1, 0.018 (95% CI: 0.016–0.020) in cluster 2, respectively. The mean gene distance is 0.040 (95% CI: 0.036–0.044) between cluster 1 and cluster 2. About 91.5% sequences have over the identity of 99%.
Estimation of positive selection sites in HuNov GII.17
The selection pressures on each nucleic acid site in the VP1 gene of GII.17 are analyzed using adaptive evolution online server (http://www.datamonkey.org/). Three positive selection sites are found (as shown in Table 1). The mean dN/dS is 0.160.
Table 1
| Amino acid change | FEL | MEME |
|---|---|---|
| Asn377Asp | √ | √ |
| Asp396Gly | √ | – |
| Glu, Pro, Val411Leu | √ | √ |
Mean dN/dS =0.160. Cutoff P value ≤0.05. Glu, glutamic acid; Pro, proline; Asn, asparagine; Asp, aspartic acid; Gly, glycine; Val, valine; Leu, leucine; HuNoV, human Norovirus.
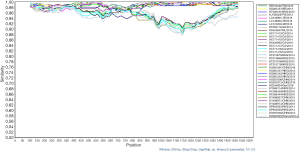
Similarity analyses of the Capsid VP1 gene in the present GII.17 strains
Similarity analysis of VP1 gene shows that the similarity of more than 93% in shell was found in shell domain while the similarity of 87% to 95% was in protruding domain (shown in Figure 2).
Discussion
In past decades, GII.4 had been the predominant genotype of HuNov leading human AGE. However, in recent years, GII.17 emerged and gradually substituted for GII.4 in HuNov infections in some countries (14). In this study, we download global VP1 full-length sequences of GII.17 from GenBank. Phylogenetic analysis showed that GII.17 circulating in present has developed into two clusters. The gene distance between cluster 1 and cluster 2 reaches to 0.040. Which is far less than the gene distance between clusters of GII.4. This suggested that the origin time of GII.17 is later than GII.4. Kobayashi et al. classified GII genotype Norovirus into three lineages (7). Of which, lineage 1 includes GII.1, GII.2, GII.5, GII.6, GII.10, GII.11, GII.12, GII.13, GII.16, GII.17, GII.18, GII.19.GII.21, GII.22. The common ancestor of lineage 1 date back to around 1819 CE (95% HPDs). Among lineage 1, the origin time of GII.17, GII.18, GII.19, GII.21, and GII.22 was later than other genotypes of lineage 1 about more than 100 years. In our study, we date GII.17 back to 1984.6 (1926.9–1995.0), which is accordance with the emergency and prevalence of GII.17 only in recently years (14,15). Moreover, compared with GII.4, the VP1 gene distance of GII.17 is much smaller than GII.4 (7). This also supports the diversity time is shorter than other genotypes of GII. We also estimated the HuNov GII.17 VP1 gene evolutionary rate as 2.31×10−3 substitutions/site/year, our observation is nearly consistent with the study by Bok et al. (2.3×10−3 substitutions/site/year) (9).
Our result also shows that the similarities of the shell domains is relatively high while that of protruding domain is lower. Previous studies showed that more epitopes of GII.17 HuNov located in protruding domain of VP1. More divergence in this domain may be associated with escaping human immunity response (16). Moreover, we found 3 positive selection sites in protruding domain of HuNov GII.17. It implied that HuNov got positive selection under human immunity pressure in vivo, and more adaptive to virus replication in vivo than before.
In conclusion, the common ancestor of GII.17 diverged from the other genotype of GII around (1926.9–1995.0) at a high evolutionary rate, although evolutionary rate of GII.17 is lower than the other genotype of GII. The protruding domain of GII.17 capsid gene had a higher divergence than the other domain of it. The existence of positive selection sites may increase the adaptivity of GII.17 living in human body, and endow it the potential of becoming the predominant GII in future.
Acknowledgments
Funding: This work was supported by Six Talent Peaks Project in Jiangsu Province (No. WSN-017) and Jiangsu Province “333” project (No. LGY2016021). Gaofeng Clinical Medicine Grant Support (No. 2017269).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jphe.2019.03.04). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ahmed SM, Hall AJ, Robinson AE, et al. Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet Infect Dis 2014;14:725-30. [Crossref] [PubMed]
- Burke RM, Shih SM, Yen C, et al. Burden of Severe Norovirus Disease in Taiwan, 2003-2013. Clin Infect Dis 2018;67:1373-8. [Crossref] [PubMed]
- Desai R, Hall AJ, Lopman BA, et al. Norovirus disease surveillance using Google Internet query share data. Clin Infect Dis 2012;55:e75-8. [Crossref] [PubMed]
- Costantini V, Morantz EK, Browne H, et al. Human Norovirus Replication in Human Intestinal Enteroids as Model to Evaluate Virus Inactivation. Emerg Infect Dis 2018;24:1453-64. [Crossref] [PubMed]
- Tsang TK, Chen TM, Longini IM Jr, et al. Transmissibility of Norovirus in Urban Versus Rural Households in a Large Community Outbreak in China. Epidemiology 2018;29:675-83. [Crossref] [PubMed]
- van Beek J, de Graaf M, Al-Hello H, et al. Molecular surveillance of norovirus, 2005-16: an epidemiological analysis of data collected from the NoroNet network. Lancet Infect Dis 2018;18:545-53. [Crossref] [PubMed]
- Kobayashi M, Matsushima Y, Motoya T, et al. Molecular evolution of the capsid gene in human norovirus genogroup II. Sci Rep 2016;6:29400. [Crossref] [PubMed]
- Bull RA, White PA. Mechanisms of GII.4 norovirus evolution. Trends Microbiol 2011;19:233-40. [Crossref] [PubMed]
- Bok K, Abente EJ, Realpe-Quintero M, et al. Evolutionary dynamics of GII.4 noroviruses over a 34-year period. J Virol 2009;83:11890-901. [Crossref] [PubMed]
- Chan MC, Lee N, Hung TN, et al. Rapid emergence and predominance of a broadly recognizing and fast-evolving norovirus GII.17 variant in late 2014. Nat Commun 2015;6:10061. [Crossref] [PubMed]
- Parra GI, Squires RB, Karangwa CK, et al. Static and Evolving Norovirus Genotypes: Implications for Epidemiology and Immunity. PLoS Pathog 2017;13:e1006136 [Crossref] [PubMed]
- Zhu S, Watanabe M, Kirkpatrick E, et al. Regulation of Norovirus Virulence by the VP1 Protruding Domain Correlates with B Cell Infection Efficiency. J Virol 2015;90:2858-67. [Crossref] [PubMed]
- Nagasawa K, Matsushima Y, Motoya T, et al. Phylogeny and Immunoreactivity of Norovirus GII.P16-GII.2, Japan, Winter 2016-17. Emerg Infect Dis 2018;24:144-8. [Crossref] [PubMed]
- Han J, Ji L, Shen Y, et al. Emergence and predominance of norovirus GII.17 in Huzhou, China, 2014-2015. Virol J 2015;12:139. [Crossref] [PubMed]
- Sang S, Yang X. Evolutionary dynamics of GII.17 norovirus. PeerJ 2018;6:e4333 [Crossref] [PubMed]
- Qian Y, Song M, Jiang X, et al. Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths. J Virol 2019;93: [Crossref] [PubMed]
Cite this article as: Zhang Y, Deng X, Chen Q, Guo H. Molecular evolution of the major capsid gene in human Norovirus GII.17. J Public Health Emerg 2019;3:6.


