
Toward tuberculosis eradication: diagnostic, resistance and monitoring insights
Tuberculosis (TB) began with civilization
TB is a highly infectious and contagious airborne disease with worldwide prevalence second only to human immunodeficiency virus (HIV) as humankind’s deadliest disease (1). It is primarily caused by the Mycobacterium tuberculosis (Mtb) which infected 9.6 million people in 2014. While no society in any part of the world is totally shielded from TB, most cases were seen in Asia (58%) and Africa (28%) (2,3). TB mortalities stood at 1.5 million out of which 400,000 suffered co-infection with HIV (3). Despite TB being declared as a universal public health emergency more than two decades ago, the number of TB deaths remain unacceptably high thus enforcing its status as a major global health problem. Controlling the TB epidemic has been hampered by many factors. Chief among these is the evolution of drug resistant strains of Mtb. Having co-existed with human hosts over many centuries, the bacteria has developed sophisticated immune escape and transmission strategies (4) and currently there exists multidrug resistant TB (MDR-TB) and extremely drug resistant TB (XDR-TB). Other factors are HIV co-infection, inadequacy of the current BCG vaccine in providing lifelong immunity and a lack of rapid, inexpensive and accurate diagnostic methods (5). Treatment of MDR-TB and XDR-TB patients requires administering a cocktail of expensive and often more toxic antibiotics for a long duration of time. Observations from clinicians treating patients with MDR-TB and XDR-TB frequently include paradoxical inflammatory episodes after weeks of response to therapy. Therefore the availability of validated biological or surrogate markers that can be used to monitor response to treatment would be extremely helpful in identifying patients at risk of treatment failures so as to personalize their treatment regimen.
TB terrorizes and the body fights back
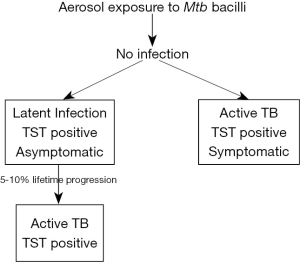
The interplay between Mtb and the host is complicated with many inadequately understood disease states. TB usually affects the lungs (pulmonary TB) resulting in severe coughing, fever, and chest pains but can affect any part of the human body (extra-pulmonary TB) (6). It is an air droplet infection acquired through inhalation of infectious aerosolized bacteria expelled from close contacts suffering from pulmonary TB. Following inhalation, some persons develop an effective immune response resulting in clearance of the mycobacteria from the lungs. A majority of individuals however develop asymptomatic latent infection where Mtb growth is inhibited but lies within them in a dormant state. These persons do not also transmit the disease. There is however a 5–10% lifetime risk of disruption of Mtb containment by the host resulting in rapid Mtb replication. This happens only in a relatively small percentage of latently-infected individuals who are then said to have active (primary) TB as shown in Figure 1 (7). Additionally, the disease may propagate from the lungs to affect other organs not limited to the brain, heart, lung, ileum, kidneys and bone (8). A startling World Health Organization statistic is that about a third of the world’s population have a latent TB infection. HIV-infected and immunocompromised persons such as those receiving treatment for cancer are at a higher risk of developing active TB (2).
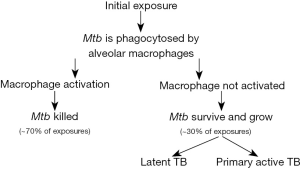
TB progresses after inhalation starting in the alveolar spaces. Mtb is an intracellular pathogen and when it encounters alveolar macrophages within the lungs, they are phagocytosed with an attempt to kill them. When the kill attempts are unsuccessful, the bacilli replicate at a rapid rate within dendritic cells and the macrophages as shown in Figure 2. Infected alveolar macrophages may remain in interstitial spaces of the lung or migrate from the alveolar space and are taken up by blood vessels or lymphatics. Other macrophages and lymphocytes are then recruited which surround the infected macrophages and begin the formation of what is known as a granuloma. The production of IL-1α, IL-1β and other host inflammatory cytokines are then triggered. This pro inflammatory response is however opposed by anti-inflammatory cytokines which aids the infection making it undetectable until after about 2–12 weeks of infection (2). Antigen presenting cells activate the adaptive immune response with the induction of both CD4+ and CD8+ T-cells. During this time there is continuous production of cytokines such as IFN-gamma and mycobacteria can acquire mutations, change expression of their genes and escape T-cell recognition (2). Between MTB infection and T-cell response, TB is repressed to an inactive form where infected individuals are asymptomatic and cannot transmit the disease. At some point in time during latency, exogenous factors can be activated which reactivates bacteria from a suppressed state with the resumption of bacterial replication (2).
TB treatment, drug resistance and paradoxical inflammatory response
There was a crucial breakthrough in the fight against TB in 1943 when streptomycin was discovered as effective against the mycobacteria. Subsequently, other equally effective drugs were introduced for use. Mtb however developed resistance to these individual drugs due to selection pressure (6). The current treatment for drug-susceptible TB involves four drugs used jointly for 2 months, followed by two drugs used in combination for 4 months (Table 1) with the aim of combating drug resistance. This treatment plan has been mostly effective with being its complexity, long duration of therapy and significant toxicity being the only drawbacks (9).
Table 1
| Drug | Daily dose | Adverse effects |
|---|---|---|
| Isoniazid (INH) | 5 mg/kg (maximum daily dose 300 mg) | Hepatitis, peripheral neuropathy, lupus-like syndrome |
| Rifampicin (RIF) | 10 mg/kg (maximum daily dose 600 mg) | Orange discoloration of secretions, hepatitis, gastrointestinal upset, fever |
| Pyrazinamide (PZA) | 25 mg/kg | Hepatitis, arthritis, hyperuricemia |
| Ethambutol (EMB) | 15 mg/kg | Optic neuritis |
TB, tuberculosis.
Global surveys conducted in the mid-1990s showed resistance by Mtb to isoniazid and rifampicin mainly due to incomplete and improper drug application. This led to the introduction and use of second-line anti-TB drugs namely fluoroquinolones, injectable aminoglycosides (kanamycin, amikacin) and cyclic peptides (capreomycin), para-aminosalicylic acid, cycloserine, prothionamide and thiacetazone. These drugs though more expensive, toxic and offering lower cure rates have also not been spared from Mtb resistance due to increased use leading to the emergence of extremely drug resistant TB (XDR-TB) (10-12). There has been little change in recent years in the number of MDR-TB cases. 3.3% of new TB cases and 20% of previously treated cases are estimated to have MDR-TB globally. An estimated number of 190, 000 people suffered deaths due to MDR-TB in 2014 and a further 9.7% of people with MDR-TB have XDR-TB (3).
Classification of drug resistance
Drug-resistant TB is a term encompassing three categories: mono resistance, multi-drug resistance and extensive drug resistance. Mono resistance refers to resistance of Mtb to one of the first-line anti-TB drugs. Multi-drug resistant TB is defined as resistance to two of the most potent first-line anti-TB drugs, rifampicin (RIF) and isoniazid (INH) either with or without resistance to any other first line anti-TB drug (3). Extensive drug resistance arises when the MDR-TB organism is resistant to any fluoroquinolone and at least one of the three second-line injectable drugs (10,13-15).
A TB-infected individual on drug treatment can develop drug resistance de novo or acquire it through infection with already resistant strains—primary resistance (12). Development of drug resistance in Mtb is due to spontaneous chromosomal mutations in genes making up drug targets of the mycobacteria thus making it functionally insensitive to anti-TB chemotherapy (13). A mutation in either the katG or inhA regulatory region accord high- and low-level resistance to isoniazid respectively (16). Rifampicin resistance arises from a point mutation in the rpo gene of the B-subunit of DNA-dependent RNA polymerase and typically follows from isoniazid resistance. Other gene mutations causing drug resistance are shown in Table 2.
Table 2
| TB drug | Gene | Likelihood of resistance |
|---|---|---|
| Isoniazid | katG | High |
| fabD | High | |
| Rifampicin | rpoB | High |
| Pyrazinamide | pncA | High |
| Ethambutol | embC | Possible |
| Fluoroquinolones | gyrA | Low |
| Aminoglycosides, streptomycin and capreomycin | rrs | High |
| Ethionamide | rpsL | High |
| ethA | Possible |
TB, tuberculosis.
An inflammatory response affiliated with TB treatment is described as a paradoxical response otherwise known as Immune Reconstitution Inflammatory Syndrome (IRIS) (4). This is generally defined as the clinical or radiological worsening of pre-existing tuberculous lesions or the development of new lesions in a patient who primarily improves with anti-TB therapy, in the absence of proof of disease relapse or the presence of another diagnosis (18,19). This response is relatively frequent; seen in 6% to 30% of TB-infected patients receiving therapy but mostly in about 10–15% of patients with a clinical diagnosis of extra pulmonary and disseminated TB (19). Clinical presentations of this paradoxical deterioration can develop anytime between 14 and 270 days with a median time of 60 days. They include fever, headache, mental confusion, focal seizures, the worsening or appearance of a pleural effusion among others. While the exact pathogenesis of this paradoxical inflammatory response is unclear, a few suggestions on its development abound. According to proponents of the immune restitution phenomenon, active TB patients overexpress T-helper 2 cells and a reduced production of interferon-gamma. Upon initiation of anti-TB chemotherapy, the mycobacterial population is reduced significantly while the pre-treatment cellular and cytokine patterns see a reversal. This abnormal immune response to the proteins released from dead bacilli may result in an inflammatory paradoxical response (18,20). The current absence of a rapid and accurate diagnostic test for this phenomenon means diagnosis can only be ascertained when other differential diagnoses such as secondary infections, insufficient anti-TB therapy and adverse reactions due to therapy are excluded.
Developments in TB diagnosis
Sputum smear microscopy, mycobacterial culture and chest radiography have been the bedrock of TB diagnosis for well over a century now. However, with the advent of near pandemic strains of MDR-TB and XDR-TB, the limitations of these methods have been laid bare. These methods are slow, costly, complicated, and laborious to execute in field conditions (21). The major diagnostic demands for TB control are detection of latent TB infection, detection of active TB and drug resistance identification. Diagnostic tests for TB are categorized either as phenotypic—bacteria from patient material (usually sputum) is inoculated into a culture medium containing the drug of interest and the appearance (indicating resistance) or absence (indicating susceptibility) of Mtb growth is detected, or genotypic—chromosomal DNA is required to detect the presence of specific mutations with known associations with drug resistance (22,23). Drug resistance in Mtb is encoded on the bacterial chromosome thus making rapid detection by molecular methods possible. This helps to address the challenge of phenotypic drug susceptibility testing which may require mycobacterial culture for a couple months at least (12). Drug Susceptibility Testing (DST) is especially more important now as inability to identify and effectively treat MDR-TB and XDR-TB patients means these patients would pass on their drug resistant TB strains to the rest of the population.
Recent developments in diagnostic technologies are founded on one of the two categories stated above. Nucleic acid (or direct) amplification tests (NATs) amplify nucleic acid regions specific to the Mtb complex and can be used on sputum. Reviews conducted on NATs show that while they have high specificity for both pulmonary and extra pulmonary TB their sensitivity is lower and highly variable across different studies. Since a negative NAT test cannot rule out the diagnosis of TB, they cannot be used with smear-negative specimens. Loop-mediated isothermal amplification assay (LAMP) works similarly to NATs and can indicate the presence of six distinct regions on the target gene. It has the advantage of speed and simplicity and has been recommended as an alternative to sputum smear microscopy in resource-poor settings. There is still insufficient evidence on its accuracy in the detection of TB compared to culture (21,22).
Immunological assays with a focus on antibody detection have also been employed in TB diagnosis. These assays have faced serious drawbacks as the proteins and genes expressed by Mtb depends on the stage of disease thus making them non-specific and incapable of distinguishing between TB, latent TB infection and nontuberculous mycobacteria. Current immune-assay based developments have thus tuned their focus to the detection of antigens, for example antigen-capture ELISA and/or circulating immune complexes rather than antibodies. This when perfected will enable specific distinction between latent and active TB (21).
Another DNA strip-based tests are line-probe assays. These rely on PCR and reverse hybridization methods to simultaneously detect Mtb and any mutations associated with drug resistance. Pai et al., write that line-probe assays have high sensitivity and specificity when culture isolates are used but lose these important characteristics when used directly on clinical specimens like sputum. Another impediment to the use of line-probe based assays is that they are expensive and can only be used in places where higher quality laboratory equipment exist. This therefore limits their use in low-income, high-burden countries (21).
Mycobacteriophage-based assays utilize bacteriophages to infect Mtb. Phage replication can only occur in samples containing viable Mtb which helps to identify the bacilli (22). Drug resistance is confirmed when Mtb. is detected in samples that contain the drug being tested e.g., rifampicin. With these assays too, they exhibit higher sensitivity and specificity when they are used for detection of rifampicin resistance in culture isolates rather than in clinical specimens (21,22).
Pyrosequencing is a sequence-based molecular method to rapidly detect mutations within specific genes linked with resistance to isoniazid, rifampicin, the fluoroquinolones and the injectable drugs.
Microscopic observation drug-susceptibility assay (MODS) is a new, promising, reliable, inexpensive tool that can quickly detect TB and drug resistance directly from clinical (sputum) specimens. It uses simple light microscopy to detect early growth of Mtb. The addition or otherwise of antimicrobial drugs to the broth medium enables its use for drug susceptibility testing (21). The MODS assay’s rapidity and simplicity make it a worthwhile tool to be optimized for testing drug resistance to second-line drugs and to detect XDR-TB especially in low resource settings (24).
The development of more sensitive molecular diagnostic tools would aid the detection of drug resistant disease before treatment begins so as to tailor chemotherapy to patient’s specific needs. In the long run this will serve the purpose of restraining the transmission of drug-resistant Mtb strains (12).
Candidate biomarkers and validation strategies
Clinicians being able to monitor TB-infected patients response to therapy is a hurdle that when overcome will help massively in the global goal of eliminating TB. Current MDR-TB and XDRTB duration of therapy is very long. In all of this time there are no valid surrogate markers available that can be measured to ascertain if a patient is responding to therapy or not. This limitation becomes more pronounced in the event of a paradoxical inflammatory response as clinicians may be forced to tinker with patient medication regimen in error.
Biological markers (or biomarkers) are measurable characteristics that indicate normal biological processes, pathogenic processes, or pharmacological responses to a medical intervention. Many biomarkers have been studied in recent times however very few have been clinically significant—that is become clinically acceptable predictors of therapeutic efficacy (25). Biomarkers may be grouped as either static or dynamic (or functional) assays. Static assays involve the measurement of levels of a peculiar substance in a clinical sample whereas dynamic (or functional) assays measure a process for example a response to an in vivo or in vitro stimulus. In monitoring disease activity in TB, an appropriate biomarker would be that which will not be masked by other attendant illnesses or therapies. A potential biomarker that can predict early remission of mycobacterial load in infected patients would be useful as such patients would receive a short course of therapy and be spared unnecessary toxic drug effects. In the same way a biomarker that would give indication of a patient being at higher relapse of reactivation risk would advise special treatment strategies on the patient in question (25).
Sputum culture status after 2 months of TB treatment has been evaluated as a candidate biomarker in monitoring disease activity. However, it was generally insensitive as only half of all relapse cases could be identified and also could not provide a verifiable positive predictive value to guide the treatment of individual patients. An interesting development is trying to figure out which mycobacteria markers can be measured in urine as that is an easy patient product to collect. A study has identified small Mtb IS6110DNA fragments termed transrenal (tr) DNA in the urine of about 80% of active TB patients but not in controls. It has the advantage of being useful in monitoring disease activity especially in children who usually have difficulty producing sputum. This method however requires PCR amplification technical support absence of which assay sensitivity may not be enough for strains with low IS6110 copy numbers. Antibody levels to some Mtb antigens e.g., alanine dehydrogenase have been explored as a potential marker to monitor disease activity at diagnosis and during treatment however none has so far exhibited results of promising value (25).
The search for biomarkers to monitor outcome of TB therapy has also extended to non-specific immune activation markers. Intercellular adhesion molecule (ICAM) 1 is a leucocyte integrin ligand principally delivered by endothelial cells. In TB patients, measures of its soluble form sICAM1 are raised at diagnosis according to extent of disease, and are lowered with response to anti-TB drugs. One study design which included a lowering of sICAM1 during the 1st week of drug therapy forecasted 2 months sputum culture conversion. Other baseline activation marker measurements with similar forecasts include serum C-reactive protein (CRP), soluble urokinase plasminogen activator receptor (suPAR), soluble tumour necrosis factor receptor (sTNFR) 1 and sTNFR2. Despite being unable to serve as treatment effect indicators, these baseline markers may give an idea of treatment relapse when associated with other recognized treatment relapse baseline markers such as bacterial burden and the presence of cavitary disease (25).
The expression of cytokines in TB could also be further studied to ascertain their application as potential biomarkers in TB disease monitoring. Interferon γ is needed for protection against mycobacterial infections. IFN-γ-inducible protein10 (IP-10/CXCL10) is a chemokine that plays a role in delayed hypersensitivity reactions. CXCL10 is assumed to be a non-specific marker of inflammation in pulmonary diseases (26). It aids in the response of Th1 cells to inflammatory targets and further attracts monocytes and activated T lymphocytes to these target areas. Pentraxin 3 (PTX3)/TNF-stimulated gene 14 (TSG-14) belongs to the pentraxin family which are involved in the acute-phase response in the event of an injury, trauma or infection. In a prospective study assessing the plasma levels of CXCL10 and/orPTX3, results showed that these two markers were both elevated in active TB patients compared to healthy controls. TB patients who were healed showed a reduction in the levels of both markers in plasma in contrast with treatment-failure patients who had persistently increasing levels of these markers in plasma. These markers could therefore serve the purpose of identifying persons at risk of disease in the case of healthy contacts or be used during disease treatment follow ups to confirm efficacy of treatment or identify treatment-failure patients early. These markers are however not as groundbreaking in the biomarker search as one may assume since they only represent the effects of long-lasting active inflammatory responses. Various external or internal stimuli and not necessarily TB disease can also cause the production of IFN-γ, TNF-α and other pro-inflammatory agents leading to their release. For high disease-burden and resource-poor nations however, these markers which are detectable by simple ELISA assay in combination with conventional clinical criteria could give an early indication of disease outcome (27).
Concluding remarks and future perspectives
There is currently a heavy clinical dependence on phenotypic DST especially in high disease burden and resource poor settings. This method is often too slow and not sufficiently validated for drugs other than the first line drugs and second line injectables to guide individual patient management. Therefore, I support calls for the WHO to be more proactive and outsource some key functions to humanitarian response groups and other global agencies that can be on the ground in all high disease burden regions (28). These bodies can provide the technical expertise such as whole genome sequencing in time to guide drug intervention. The recent outbreak of drug-resistant TB in Papua New Guinea and the slow WHO response emphasizes this need. Also, with emerging technological advancements in diagnostic equipment, global efforts should be aimed at subsidizing the cost of these equipments to all countries around the world where none exist. Ultimately, the discovery of new classes of anti-TB drugs, biomarkers and validated surrogate endpoints hold the greatest promise in disease eradication. Currently, there is no biomarker that has achieved clinical significance for disease monitoring during treatment. It is possible that there is no one single biomarker that can serve this purpose. Future research should look at multiple biomarkers that account for the complex means of disease development and mechanism of actions of all currently available medications. The discovery of biological markers and surrogate endpoints that will differentiate active TB patients from healthy individuals across diverse population groups, return to normal levels with drug treatment and reproducibly predict clinical outcomes would be a major breakthrough in TB control.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jphe.2016.12.12). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rahman A, Sahrin M, Afrin S, et al. Comparison of Xpert MTB/RIF Assay and GenoType MTBDRplus DNA Probes for Detection of Mutations Associated with Rifampicin Resistance in Mycobacterium tuberculosis. PLoS One 2016;11:e0152694 [Crossref] [PubMed]
- Fogel N. Tuberculosis: a disease without boundaries. Tuberculosis (Edinb) 2015;95:527-31. [Crossref] [PubMed]
- World Health Organization. Global Tuberculosis Report (2015). WHO/HTM/TB/2015.22
- Ralph AP, Anstey NM, Kelly PM. Tuberculosis into the 2010s: is the glass half full? Clin Infect Dis 2009;49:574-83. [Crossref] [PubMed]
- John SH, Kenneth J, Gandhe AS. Host biomarkers of clinical relevance in tuberculosis: review of gene and protein expression studies. Biomarkers 2012;17:1-8. [Crossref] [PubMed]
- Majewski K, Rybczyńska M, Wódz K. Evaluation of detection and drug resistance of Mycobacterium tuberculosis in patients in the Łódzkie voivodship in 2009-2013. Przegl Epidemiol 2015;69:453-8, 575-80. [PubMed]
- Sridhar S, Pollock K, Lalvani A. Redefining latent tuberculosis. Future Microbiol 2011;6:1021-35. [Crossref] [PubMed]
- Johnson DH, Via LE, Kim P, et al. Nuclear imaging: a powerful novel approach for tuberculosis. Nucl Med Biol 2014;41:777-84. [Crossref] [PubMed]
- Wong EB, Cohen KA, Bishai WR. Rising to the challenge: new therapies for tuberculosis. Trends Microbiol 2013;21:493-501. [Crossref] [PubMed]
- Chakroborty A. Drug-resistant tuberculosis: an insurmountable epidemic? Inflammopharmacology 2011;19:131-7. [Crossref] [PubMed]
- Ajbani K, Shetty A, Mehta A, et al. Rapid diagnosis of extensively drug-resistant tuberculosis by use of a reverse line blot hybridization assay. J Clin Microbiol 2011;49:2546-51. [Crossref] [PubMed]
- Müller B, Borrell S, Rose G, et al. The heterogeneous evolution of multidrug-resistant Mycobacterium tuberculosis. Trends Genet 2013;29:160-9. [Crossref] [PubMed]
- Porvaznik I, Mokry J, Solovic I. Classical against molecular-genetic methods for susceptibility testing of antituberculotics. Adv Exp Med Biol 2015;835:15-22. [Crossref] [PubMed]
- Bonnet M, Pardini M, Meacci F, et al. Treatment of tuberculosis in a region with high drug resistance: outcomes, drug resistance amplification and re-infection. PLoS One 2011;6:e23081 [Crossref] [PubMed]
- Jeon D. Medical Management of Drug-Resistant Tuberculosis. Tuberc Respir Dis (Seoul) 2015;78:168-74. [Crossref] [PubMed]
- Ahmad S, Mokaddas E. Current status and future trends in the diagnosis and treatment of drug-susceptible and multidrug-resistant tuberculosis. J Infect Public Health 2014;7:75-91. [Crossref] [PubMed]
- Outhred AC, Jelfs P, Suliman B, et al. Added value of whole-genome sequencing for management of highly drug-resistant TB. J Antimicrob Chemother 2015;70:1198-202. [PubMed]
- Fernández-Fúnez A. Paradoxical response during anti-tuberculosis treatment in immunocompetent patients. Med Clin (Barc) 2009;133:637-43. [PubMed]
- Brown CS, Smith CJ, Breen RA, et al. Determinants of treatment-related paradoxical reactions during anti-tuberculosis therapy: a case control study. BMC Infectious Diseases 2016;16:479. [Crossref] [PubMed]
- Cheng VC. Paradoxical Response during Anti-tuberculosis Therapy. The Hong Kong Medical Diary. Vol.11 No.1 January 2006.
- Pai M, Kalantri S, Dheda K. New tools and emerging technologies for the diagnosis of tuberculosis: part II. Active tuberculosis and drug resistance. Expert Rev Mol Diagn 2006;6:423-32. [Crossref] [PubMed]
- Grandjean L, Moore DA. Tuberculosis in the developing world: recent advances in diagnosis with special consideration of extensively drug-resistant tuberculosis. Curr Opin Infect Dis 2008;21:454-61. [Crossref] [PubMed]
- Theron G, Peter J, Richardson M, et al. The diagnostic accuracy of the GenoType(®) MTBDRsl assay for the detection of resistance to second-line anti-tuberculosis drugs. Cochrane Database Syst Rev 2014;CD010705 [PubMed]
- Hillery N, Groessl EJ, Trollip A, et al. The Global Consortium for Drug-resistant Tuberculosis Diagnostics (GCDD): design of a multi-site, head-to-head study of three rapid tests to detect extensively drug-resistant tuberculosis. Trials 2014;15:434. [Crossref] [PubMed]
- Wallis RS, Doherty TM, Onyebujoh P, et al. Biomarkers for tuberculosis disease activity, cure, and relapse. Lancet Infect Dis 2009;9:162-72. [Crossref] [PubMed]
- Hasan Z, Rao N, Salahuddin N, et al. Mycobacterium tuberculosis Sonicate-Induced IFNγ, CXCL10 and IL10 can Differentiate Severity in Tuberculosis. Scand J Immunol 2012;75:220-6. [Crossref] [PubMed]
- Azzurri A, Sow OY, Amedei A, et al. IFN-gamma-inducible protein 10 and pentraxin 3 plasma levels are tools for monitoring inflammation and disease activity in Mycobacterium tuberculosis infection. Microbes Infect 2005;7:1-8. [Crossref] [PubMed]
- Negin J, Dhillon RS. Outsourcing: how to reform WHO for the 21st century. BMJ Glob Health 2016;1:e000047 [Crossref]
Cite this article as: Sarpong KA. Toward tuberculosis eradication: diagnostic, resistance and monitoring insights. J Public Health Emerg 2017;1:12.


