
Plasma microRNA expression in workers occupationally exposed to benzene
Introduction
Benzene is a confirmed organic carcinogen, and one of the most common environmental pollutants. However, it is still widely used as an industrial solvent, representing a significant occupational hazard. Long-term exposure to benzene may lead to a range of hematologic disorders such as aplastic anemia (1), acute myelogenous leukemia, and other forms of leukemia (2). The adverse effects of benzene exposure are probably mediated through its metabolites via multiple pathways, including oxidative stress, DNA damage, cell cycle regulation and programmed cell death (3). To date, the mechanism underlying benzene-induced hematotoxicity and leukemogenicity has not been fully elucidated.
It was previously reported that exposure to benzene or its metabolites was associated with the alterations in the expression patterns of genes that were relevant to the toxic effects of benzene (4,5). Environmental exposures might alter gene expression by affecting the expression levels of microRNA (miRNA) that function as an important gene regulator (6,7). Moreover, a great deal of research shows that exposure to hazardous environmental factors may notably disrupt miRNA expression patterns (8-10). Despite intensive studies on benzene over several decades, investigations on plasma miRNA expression in benzene-exposed workers are lagging behind.
Circulating miRNA has attractive interests as a non-invasive biomarker of physiological and pathological conditions. In a previous study (11), we have investigated the potential effects of chronic benzene poisoning (CBP) and benzene exposure on miRNA expression, and identify CBP-related miRNAs. In the present study, we further identify plasma miRNAs that are associated with benzene exposures with a larger sample sizes.
Methods
Subjects
In the present study, we enrolled 118 healthy benzene-exposed workers who were employed at the work posts of benzene for at least one year. Meanwhile, other 91 subjects without occupational benzene exposure were enrolled as controls. After participants provided informed consent, we administered a questionnaire to collect information on demographic characteristics, lifetime occupational history, lifestyle habits (smoking and alcohol consumption), disease history, environmental benzene exposures, history of hematotoxicity drug utilization, physical and chemical factor exposure. Subjects with previous or present treatment on hematotoxicity drugs (e.g., chloromycetin, sulfonamides), habitual exposure to other chemical or physical factors causing hematotoxicity (e.g., ionizing radiation, arsenic) and recent infection were excluded from the study. Each worker donated approximately 5 mL fasting venous blood (EDTA-anticoagulated), and the plasma was isolated within one hour and stored at −80 °C until RNA extraction. Exposure estimation was based on benzene concentration monitoring data at the work place provided by Jiangsu Provincial Center for Disease Prevention and Control. This project was approved by the Ethics Committee of Jiangsu Provincial Center for Disease Prevention and Control, and ethics were respected throughout the whole study period.
Study design
According to whether exposed to benzene, subjects were classified into two groups: exposure (118 healthy benzene-exposed individuals) and control (91 healthy non-benzene exposed individuals).In the discovery stage, we selected 10 workers from the exposed group, and 10 matched subjects from the control group. We intentionally frequency-matched the distribution of important general characteristics, including sex, age (±5 years), nationality (Han Chinese), smoking status, drinking status, between these two groups to minimize their confounding effects on miRNA expression profiles. We prepared a 2 mL pooled plasma sample for each group that included 200 µL of plasma from each subject. We then subjected two plasma pools to miRNA microarray analysis and compared miRNA expression profiles between these two groups. To focus on the most likely related miRNAs in the validation stage, we selected candidate miRNAs from the microarray analysis based on the following criteria: demonstrating at least 1.0 fold higher or −1.0 fold lower expression between any two out of three groups, expressing more than 500 copies in any group, and giving priority to choosing molecules that may be associated with a benzene response based on the literature. These 20 subjects in the discovery stage were also included in the validation stage.
RNA isolation
We isolated total RNA from the two EDTA-anticoagulated plasma pools collected in the discovery stage for microarray analysis; we also isolated total RNA from200 µL EDTA-anticoagulated plasma from each subject in the validation stage, which was analyzed by qRT-PCR (Norgen’s Total RNA Purification Kit, Norgen Biotech, Thorold, Canada). For RNA isolation in the validation stage, we added caenorhabditis elegans miRNA [cel-miR-238 (25 fmol), Takara, Dalian, China] as an internal control prior to the isolation procedure.
MicroRNA (miRNA) microarray assay
miRNA expression profiles of two plasma pools were examined using the microarray assay (version 19.0) performed by service provider (subsidiary of LC Sciences, Hangzhou, China) as described previously (12). Briefly, about 0.5 µg total RNA samples from plasma were 3'-extended with a poly(A) tail, to which an oligonucleotide tag was ligated for later fluorescent dye staining. On the microfluidic chip, each detection probe consists of a chemically modified nucleotide coding segment complementary to target microRNA or other RNA (control sequences) and a spacer segment of polyethylene glycol to extend the coding segment away from the substrate. After hybridization, tag-conjugating Cy3 dye was circulated through the microfluidic chip for dye staining. Microarray was then scanned with a laser scanner (GenePix 4000B, Molecular Device), and raw data were imput into Array-Pro image analysis software (Media Cybernetics). The data were analyzed by firstly subtracting the background, and then normalizing the signals using a LOWESS filter (Locally-weighted Regression) (13). The signal values, higher than the background value by three folds of standard deviation, were considered valuable. The fold changed values of any two out of three sets of detected signals (log2 transformed and balanced) were calculated.
Real-time quantitative PCR assay
RNAs were reverse transcribed (TaqMan® miRNA Reverse Transcription Kit) following the manufacturer’s protocol and then subjected to real time PCR in triplicate using the TaqMan miRNA Assay Kit and an ABI PRISM® 7900HT Real-Time PCR System (all from Applied Biosystems, Foster City, CA). The expression level of each miRNA was individually normalized to cel-miR-238.
Statistical analysis
The ratios of two sets (exposure and control) from array analysis were calculated (as log2 value). We evaluated the differences of general characteristics between different groups (control vs. exposed groups) by Student’s t-test (for continuous variables) and chi-square test (for categorical variables). In the validation stage, the relative expression level of each miRNA in an individual sample was calculated according to the equation 2−∆Ct, in which ∆Ct = Ct miRNA − Ct cel-miR-238. Then the relative quantification value underwent a Log2-transformation to compare the expression levels of candidate miRNAs between groups. Statistical analysis was performed with SPSS software and two-sided P<0.05was considered statistically significant.
Results
Subject characteristics
A total of 209 participants were enrolled in our study, including 91 non-exposure controls (64 men and 27 women, mean age: 37.6±9.7), 118 healthy exposure subjects (79 man and 39 women, mean age: 37.3±8.2), of which 10 participants from each group were selected for the microarray analysis. The detailed information about research subjects was listed in Table 1. In this study, distributions of sex, age, smoking and drinking status did not significantly different (all P>0.05) between the exposure and control.
Table 1
| Variable | Discovery stage | Validation stage | |||||
|---|---|---|---|---|---|---|---|
| Controls (n=10) | Exposed (n=10) | P value | Controls (n=91) | Exposed (n=118) | P value | ||
| Sex (male/female) | 7/3 | 7/3 | 1.000 | 64/27 | 79/39 | 0.654 | |
| Age (mean ± SD) | 39.0±4.0 | 39.1±4.7 | 0.959 | 37.6±9.7 | 37.3±8.2 | 0.809 | |
| Smoking (n, %) | 4 (40.0) | 4 (40.0) | 1.000 | 33 (39.7) | 50 (42.4) | 0.395 | |
| Drinking (n, %) | 2 (20.0) | 2 (20.0) | 1.000 | 32 (35.2) | 48 (40.7) | 0.474 | |
| Exposure years | − | 11.1±4.5 | − | − | 10.3±7.7 | − | |
| Exposure intensity (mg/m3) | − | 12.1±9.3 | − | − | 11.7±9.0 | − | |
MicroRNA (miRNA) microarray analysis
We carried out a miRNA microarray assay to evaluate the expression levels of 2,019 known human miRNAs listed in Sanger miRBase Release 19.0 (http://www.sanger.ac.uk/Software/Rfam/mirna/). In the discovery stage, 287 and 272 miRNAs were found to be expressed in the plasma pool of the controls and the healthy exposure subjects, respectively, and 1,171 out of 2,019 miRNAs had not been found. Further analysis revealed that 251 miRNAs were commonly detected in both two groups with the remaining 36 and 21 molecules exclusively expressed in exposures and controls, respectively (Figure 1). Notably, compared with the expression levels in the control group, 5 miRNAs demonstrated at least a 1-fold higher expression, and 12 miRNAs showed at least a −1-fold lower expression in the exposure group (Table 2). Based on the miRNA selection criteria (see “Materials and Methods”), we selected 1 significantly differentially expressed miRNAs (hsa-miR-638) from these 5 up-regulated miRNAs at the stage of validation. We also selected 2 miRNA (hsa-miR-221-3p and hsa-miR-122-5p) from these 12 down-regulated miRNAs. The expression levels and the related functions of three candidate miRNAs are shown in Table 3 (11,14-18).
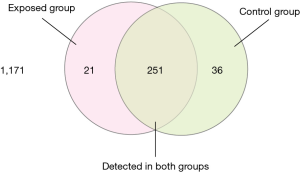
Table 2
| miRNA | Exposure | Control | Fold-change [log2(exposure/control)] |
|---|---|---|---|
| hsa-miR-652-3p | 399 | 81 | 2.30 |
| hsa-miR-30c-1-3p | 429 | 191 | 1.16 |
| hsa-miR-4301 | 143 | 65 | 1.13 |
| hsa-miR-106b-3p | 58 | 27 | 1.11 |
| hsa-miR-638 | 2,164 | 1,078 | 1.01 |
| hsa-miR-4687-3p | 53 | 107 | −1.00 |
| hsa-miR-221-3p | 633 | 1,272 | −1.01 |
| hsa-miR-122-5p | 1,124 | 2,278 | −1.02 |
| hsa-miR-6088 | 58 | 119 | −1.03 |
| hsa-miR-4632-5p | 18 | 37 | −1.03 |
| hsa-miR-3162-5p | 28 | 66 | −1.26 |
| hsa-miR-4513 | 14 | 35 | −1.30 |
| hsa-miR-4281 | 344 | 850 | −1.31 |
| hsa-miR-4433-3p | 25 | 62 | −1.33 |
| hsa-miR-6073 | 236 | 632 | −1.42 |
| hsa-miR-4655-3p | 14 | 42 | −1.59 |
| hsa-miR-505-5p | 12 | 38 | −1.72 |
Table 3
| miRNA | Discovery stage | FCa | Validation stageb | Relevant functions | Sources | |
|---|---|---|---|---|---|---|
| Exposure | Control | |||||
| hsa-miR-638 | 2,164 | 1,078 | 1.01 | 1.984 (1.020, 4.086) | CBP-related miRNAs; Benzo(a)pyrene response; regulation of differentiation and proliferation in leukemic cells | (11,14,15) |
| hsa-miR-221-3p | 633 | 1,272 | −1.01 | 0.201 (0.145, 0.289) | CBP-related miRNAs; benzene exposure response; low-level hydroquinone response | (11,16,17) |
| hsa-miR-122-5p | 1,124 | 2,278 | −1.02 | 0.378 (0.194, 0.523) | CBP-related miRNAs; Heme oxygenase-1 regulation | (11,18) |
a, the fold change value underwent a Log2-transformation; b, values are median (25th percentile, 75th percentile) qRT-PCR expression relative to cel-miR-238.
Identification of candidate microRNA (miRNA) by real-time quantitative PCR
For further exploring the possible influence of benzene exposure on miRNA expression, three miRNAs (miR-638, miR-221-3p and miR-122-5p) were selected for the expanded validation by qRT-PCR in 118 healthy exposure subjects and 91 non-exposure controls. Among those, 10 healthy exposure subjects and 10 non-exposed controls were repeated in microarray analysis stage.
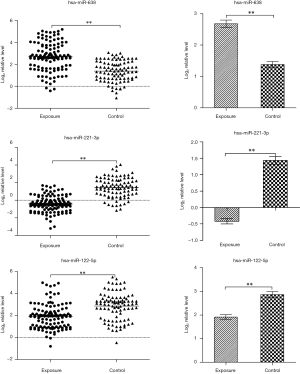
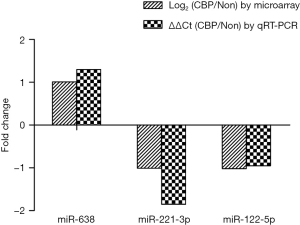
The plasma levels of the three miRNAs are presented in Figure 2. Compared with the controls, the plasma level of miR-638 was significantly up-regulated (2.68±0.12 vs. 1.38±0.10, P<0.01) while the plasma levels of miR-221-3p and miR-122-5p were significantly down-regulated (−0.42±0.08 vs. 1.44±0.12 formiR-221-3pand 1.91±0.10 vs. 2.86±0.13 for miR-122-5p, P<0.01) in the exposures. The validation results of the three miRNAs were in concordance with the microarray analysis (Figure 3).
Discussion
During the benzene related production, benzene was usually inhaled by people as its steamy form. In the liver, benzene is metabolized to phenol, which is further hydroxylated to hydroquinone (HQ), catechol, and 1,2,4-benzenetriol (2,19). HQ and catechol are oxidized by myeloperoxidase (MPO) to 1,4-benzoquinone (BQ). It was previously reported that benzene exposure was associated with mRNA expression levels (20), mitochondrial DNA copy number (21), and DNA methylation status (22) in benzene-exposed workers. Deng et al. (23) identified that dysregulated miRNAs were involved in responses to polycyclic aromatic hydrocarbons (PAHs) in exposed workers and further studies (24) revealed that miRNAs and their interactions with environmental factors might affect the effects of PAHs exposure on oxidative DNA damage and lipid peroxidation. In present study, we explored the potential effects of occupational benzene exposure on plasma miRNA expression in Chinese occupational population.
In the present study, we first employed miRNA microarray assay to examine the differential profiles of plasma miRNA expression between benzene exposure workers and non-exposed controls. In a detailed validation study, we identified three miRNAs that could be associated with the responses to benzene, including one (miR-638) that was positively associated with benzene exposure levels and two (miR-221-3p and miR-122-5p) that was negatively associated with benzene exposure levels. Previous studies have demonstrated that exposure to hazardous environmental factors may significantly disrupt miRNA expression patterns (8-10). These results again provided novel information about associations of benzene exposures with plasma miRNA expression in exposed workers.
Our microarray analysis and subsequent validation identified three indicated miRNAs in plasma that may affect the responses to benzene exposure. And the three miRNAs have also been reported to regulate genes that could influence the responses to benzene exposures. In the present study, miR-638 was up-regulated in the plasma of the benzene exposed subjects. The results were in line with previous studies shown that miR-638 was upregulated in immortalized human bronchial epithelial (HBE/HBER) cells exposed to BaP and in peripheral lymphocytes of PAHs-exposed (PE) workers (14). Actually miR-638 was proposed as an important regulator of development, tumorigenesis, hematopoiesis, and leukemogenesis. Indeed, miR-638 was involved in BaP-induced carcinogenesis by targeting BRCA1 (14), regulates proliferation and myeloid differentiation by targeting CDK2 (15), and highly expressed in human vascular smooth muscle cells, promoting proliferation and migration by targeting NOR1 (25). In our previous study (11), miR-638 was also up-regulated in the plasma of the benzene exposed subjects and exhibited a dual function in response to benzene depending upon the state of pathological process.
In the present study, miR-221-3p and miR-122-5p were both down-regulated in the plasma of the benzene exposed subjects. Recently, miR-221, which has been suggested to be an oncogene, was frequently reported to be significantly deregulated in blood cells of patients with leukemia (26-28). An epidemiological investigation indicated that benzene exposure may be related to elevated miR-221 expression in peripheral blood lymphocytes of petrol station attendants (17). Interestingly, other functional experiments have indicated that low-level hydroquinone, an important benzene metabolite, could inhibit miR-221 expression and cell apoptosis, while promote cell growth in TK 6 lymphoblastoid cell model (16). It could be due to the possibility that the expression of miRNA is highly specific for cell, tissue or developmental stage. This could be an example of tissue- or developmental stage-specific miRNA activation. Furthermore, miR-122 was a liver-specific miRNA and down-regulation of miR-122 was an early event in hepatocarcinogenesis (29). Marrone et al. reported that the expression levels of miR-122 was reduced in HepaRG cells treated with AFB1, a human liver carcinogen, and the AFB1-induced down-regulation of miR-122 was associated with an inhibition of the HNF4A/miR-122 regulatory pathway (30).
There were several limitations in our study: (I) our study is a cross-sectional study, which does not permit an ultimate confirmation of the association between benzene exposure and changes in three indicated miRNAs; (II) the sample sizes of exposure and control groups were relatively small and thus those subjects might been not representative. In addition, we validated only a subset of the miRNAs found in discovery stage, other miRNAs should be further validated.
Conclusions
In conclusion, our investigation suggested that benzene exposure might be related to elevated expression of miR-638 and decreased expression of miR-221-3p and miR-122-5p in human plasma.
Acknowledgments
Funding: This study was funded by Outstanding Medical Academic Leaders program of Jiangsu Province (LJ201130).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jphe.2016.12.03). XC serves as an unpaid editorial board member of Journal of Public Health and Emergency from Jan 2017 to Dec 2019. BZ serves as an Editor-in-Chief of Journal of Public Health and Emergency from Jan 2017 to Dec 2022. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This project was approved by the Ethics Committee of Jiangsu Provincial Center for Disease Prevention and Control, and ethics were respected throughout the whole study period. Informed consent was taken from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Aksoy M. Hematotoxicity and carcinogenicity of benzene. Environ Health Perspect 1989;82:193-7. [Crossref] [PubMed]
- Atkinson TJ. A review of the role of benzene metabolites and mechanisms in malignant transformation: summative evidence for a lack of research in nonmyelogenous cancer types. Int J Hyg Environ Health 2009;212:1-10. [Crossref] [PubMed]
- Sarma SN, Kim YJ, Ryu JC. Differential gene expression profiles of human leukemia cell lines exposed to benzene and its metabolites. Environ Toxicol Pharmacol 2011;32:285-95. [Crossref] [PubMed]
- Byeon SE, Yu T, Yang Y, et al. Hydroquinone regulates hemeoxygenase-1 expression via modulation of Src kinase activity through thiolation of cysteine residues. Free Radic Biol Med 2013;57:105-18. [Crossref] [PubMed]
- Huang J, Zhao M, Li X, et al. The Cytotoxic Effect of the Benzene Metabolite Hydroquinone is Mediated by the Modulation of MDR1 Expression via the NF-κB Signaling Pathway. Cell Physiol Biochem 2015;37:592-602. [Crossref] [PubMed]
- Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet 2007;8:253-62. [Crossref] [PubMed]
- Schembri F, Sridhar S, Perdomo C, et al. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc Natl Acad Sci U S A 2009;106:2319-24. [Crossref] [PubMed]
- Jardim MJ, Fry RC, Jaspers I, et al. Disruption of microRNA expression in human airway cells by diesel exhaust particles is linked to tumorigenesis-associated pathways. Environ Health Perspect 2009;117:1745-51. [PubMed]
- Bollati V, Marinelli B, Apostoli P, et al. Exposure to metal-rich particulate matter modifies the expression of candidate microRNAs in peripheral blood leukocytes. Environ Health Perspect 2010;118:763-8. [Crossref] [PubMed]
- De Flora S, Balansky R, D'Agostini F, et al. Smoke-induced microRNA and related proteome alterations. Modulation by chemopreventive agents. Int J Cancer 2012;131:2763-73. [Crossref] [PubMed]
- Liu Y, Chen X, Bian Q, et al. Analysis of plasma microRNA expression profiles in a Chinese population occupationally exposed to benzene and in a population with chronic benzene poisoning. J Thorac Dis 2016;8:403-14. [Crossref] [PubMed]
- Wang H, Zheng Y, Wang G, et al. Identification of microRNA and bioinformatics target gene analysis in beef cattle intramuscular fat and subcutaneous fat. Mol Biosyst 2013;9:2154-62. [Crossref] [PubMed]
- Bolstad BM, Irizarry RA, Astrand M, et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003;19:185-93. [Crossref] [PubMed]
- Li D, Wang Q, Liu C, et al. Aberrant expression of miR-638 contributes to benzo(a)pyrene-induced human cell transformation. Toxicol Sci 2012;125:382-91. [Crossref] [PubMed]
- Lin Y, Li D, Liang Q, et al. miR-638 regulates differentiation and proliferation in leukemic cells by targeting cyclin-dependent kinase 2. J Biol Chem 2015;290:1818-28. [Crossref] [PubMed]
- Liu LH, Liang XH, Ling XX, et al. Influences of biological characteristics and the expression of milt-221 induce by low-level hydroquinone exposure in TK6 lymphoblastoid cells. China Occupational Medicine 2011;38:102-5.
- Hu D, Peng X, Liu Y, et al. Overexpression of miR-221 in peripheral blood lymphocytes in petrol station attendants: A population based cross-sectional study in southern China. Chemosphere 2016;149:8-13. [Crossref] [PubMed]
- Izzotti A, Calin GA, Arrigo P, et al. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J 2009;23:806-12. [Crossref] [PubMed]
- Snyder R. Xenobiotic metabolism and the mechanism(s) of benzene toxicity. Drug Metab Rev 2004;36:531-47. [Crossref] [PubMed]
- Wang Q, Ye R, Ye YJ, et al. mRNA expression levels among cell regulatory and DNA damage genes in benzene-exposed workers in China. J Occup Environ Med 2012;54:1467-70. [Crossref] [PubMed]
- Shen M, Zhang L, Bonner MR, et al. Association between mitochondrial DNA copy number, blood cell counts, and occupational benzene exposure. Environ Mol Mutagen 2008;49:453-7. [Crossref] [PubMed]
- Bollati V, Baccarelli A, Hou L, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res 2007;67:876-80. [Crossref] [PubMed]
- Deng Q, Huang S, Zhang X, et al. Plasma microRNA expression and micronuclei frequency in workers exposed to polycyclic aromatic hydrocarbons. Environ Health Perspect 2014;122:719-25. [PubMed]
- Deng Q, Dai X, Guo H, et al. Polycyclic aromatic hydrocarbons-associated microRNAs and their interactions with the environment: influences on oxidative DNA damage and lipid peroxidation in coke oven workers. Environ Sci Technol 2014;48:4120-8. [Crossref] [PubMed]
- Li P, Liu Y, Yi B, et al. MicroRNA-638 is highly expressed in human vascular smooth muscle cells and inhibits PDGF-BB-induced cell proliferation and migration through targeting orphan nuclear receptor NOR1. Cardiovasc Res 2013;99:185-93. [Crossref] [PubMed]
- Undi RB, Kandi R, Gutti RK. MicroRNAs as Haematopoiesis Regulators. Adv Hematol 2013;2013:695754.
- Gimenes-Teixeira HL, Lucena-Araujo AR, Dos Santos GA, et al. Increased expression of miR-221 is associated with shorter overall survival in T-cell acute lymphoid leukemia. Exp Hematol Oncol 2013;2:10. [Crossref] [PubMed]
- Coskun E, Neumann M, Schlee C, et al. MicroRNA profiling reveals aberrant microRNA expression in adult ETP-ALL and functional studies implicate a role for miR-222 in acute leukemia. Leuk Res 2013;37:647-56. [Crossref] [PubMed]
- Takaki Y, Saito Y, Takasugi A, et al. Silencing of microRNA-122 is an early event during hepatocarcinogenesis from non-alcoholic steatohepatitis. Cancer Sci 2014;105:1254-60. [Crossref] [PubMed]
- Marrone AK, Tryndyak V, Beland FA, et al. MicroRNA Responses to the Genotoxic Carcinogens Aflatoxin B1 and Benzo[a]pyrene in Human HepaRG Cells. Toxicol Sci 2016;149:496-502. [Crossref] [PubMed]
Cite this article as: Liu Y, Chen X, Shi Y, Zhang J, Zhang H, Zhang F, Zhu B. Plasma microRNA expression in workers occupationally exposed to benzene. J Public Health Emerg 2017;1:11.


